data
Autogenerated data summary from dataMaid
Data report overview
The dataset examined has the following dimensions:
| Feature | Result |
|---|---|
| Number of observations | 4559 |
| Number of variables | 106 |
Checks performed
The following variable checks were performed, depending on the data type of each variable:
| character | factor | labelled | haven labelled | numeric | integer | logical | Date | |
|---|---|---|---|---|---|---|---|---|
| Identify miscoded missing values | × | × | × | × | × | × | × | |
| Identify prefixed and suffixed whitespace | × | × | × | × | ||||
| Identify levels with < 6 obs. | × | × | × | × | ||||
| Identify case issues | × | × | × | × | ||||
| Identify misclassified numeric or integer variables | × | × | × | × | ||||
| Identify outliers | × | × | × |
Please note that all numerical values in the following have been rounded to 2 decimals.
Summary table
| Variable class | # unique values | Missing observations | Any problems? | |
|---|---|---|---|---|
| icustay_id…1 | numeric | 4559 | 0.00 % | |
| hadm_id…2 | numeric | 4559 | 0.00 % | |
| intime | character | 4559 | 0.00 % | × |
| outtime | character | 4559 | 0.00 % | × |
| dbsource | character | 1 | 0.00 % | × |
| suspected_infection_time_poe | character | 4558 | 0.00 % | × |
| suspected_infection_time_poe_days | numeric | 4370 | 0.00 % | × |
| specimen_poe | character | 35 | 0.00 % | × |
| positiveculture_poe | numeric | 2 | 0.00 % | |
| antibiotic_time_poe | character | 4274 | 0.00 % | × |
| blood_culture_time | character | 4558 | 0.00 % | × |
| blood_culture_positive | numeric | 2 | 0.00 % | |
| age | numeric | 3831 | 0.00 % | |
| gender | character | 2 | 0.00 % | |
| is_male | numeric | 2 | 0.00 % | |
| ethnicity | character | 41 | 0.00 % | × |
| race_white | numeric | 2 | 0.00 % | |
| race_black | numeric | 2 | 0.00 % | |
| race_hispanic | numeric | 2 | 0.00 % | |
| race_other | numeric | 2 | 0.00 % | |
| metastatic_cancer | numeric | 2 | 0.00 % | |
| diabetes | numeric | 2 | 0.00 % | |
| first_service | character | 13 | 0.00 % | |
| hospital_expire_flag | numeric | 2 | 0.00 % | |
| thirtyday_expire_flag | numeric | 2 | 0.00 % | |
| icu_los | numeric | 4401 | 0.00 % | × |
| hosp_los | numeric | 4191 | 0.00 % | × |
| sepsis_angus | numeric | 2 | 0.00 % | |
| sepsis_martin | numeric | 2 | 0.00 % | |
| sepsis_explicit | numeric | 2 | 0.00 % | |
| septic_shock_explicit | numeric | 2 | 0.00 % | |
| severe_sepsis_explicit | numeric | 2 | 0.00 % | |
| sepsis_nqf | numeric | 2 | 0.00 % | |
| sepsis_cdc | numeric | 2 | 0.00 % | |
| sepsis_cdc_simple | numeric | 2 | 0.00 % | |
| elixhauser_hospital | numeric | 49 | 0.00 % | × |
| vent | numeric | 2 | 0.00 % | |
| sofa | numeric | 20 | 0.00 % | × |
| lods | numeric | 21 | 0.00 % | × |
| sirs | numeric | 5 | 0.00 % | |
| qsofa | numeric | 4 | 0.00 % | |
| qsofa_sysbp_score | numeric | 3 | 0.15 % | |
| qsofa_gcs_score | numeric | 3 | 0.07 % | |
| qsofa_resprate_score | numeric | 3 | 0.02 % | |
| aniongap_min | numeric | 35 | 0.31 % | × |
| aniongap_max | numeric | 44 | 0.31 % | × |
| bicarbonate_min | numeric | 40 | 0.07 % | × |
| bicarbonate_max | numeric | 43 | 0.07 % | × |
| creatinine_min | numeric | 98 | 0.04 % | × |
| creatinine_max | numeric | 123 | 0.04 % | × |
| chloride_min | numeric | 67 | 0.02 % | × |
| chloride_max | numeric | 65 | 0.02 % | × |
| glucose_min | numeric | 257 | 0.00 % | × |
| glucose_max | numeric | 473 | 0.00 % | × |
| hematocrit_min | numeric | 341 | 0.07 % | × |
| hematocrit_max | numeric | 331 | 0.07 % | × |
| hemoglobin_min | numeric | 139 | 0.07 % | × |
| hemoglobin_max | numeric | 130 | 0.07 % | × |
| lactate_min | numeric | 97 | 0.00 % | × |
| lactate_max | numeric | 165 | 0.00 % | × |
| lactate_mean | numeric | 209 | 0.00 % | × |
| platelet_min | numeric | 528 | 0.13 % | × |
| platelet_max | numeric | 609 | 0.13 % | × |
| potassium_min | numeric | 51 | 0.00 % | × |
| potassium_max | numeric | 73 | 0.00 % | × |
| inr_min | numeric | 69 | 5.92 % | × |
| inr_max | numeric | 99 | 5.92 % | × |
| sodium_min | numeric | 61 | 0.00 % | × |
| sodium_max | numeric | 63 | 0.00 % | × |
| bun_min | numeric | 140 | 0.04 % | × |
| bun_max | numeric | 157 | 0.04 % | × |
| bun_mean | numeric | 256 | 0.00 % | × |
| wbc_min | numeric | 349 | 0.11 % | × |
| wbc_max | numeric | 436 | 0.11 % | × |
| wbc_mean | numeric | 658 | 0.00 % | × |
| heartrate_min | numeric | 114 | 0.00 % | × |
| heartrate_max | numeric | 131 | 0.00 % | × |
| heartrate_mean | numeric | 3902 | 0.00 % | × |
| sysbp_min | numeric | 135 | 0.18 % | × |
| sysbp_max | numeric | 152 | 0.18 % | × |
| sysbp_mean | numeric | 3868 | 0.18 % | × |
| diasbp_min | numeric | 81 | 0.18 % | × |
| diasbp_max | numeric | 137 | 0.18 % | × |
| diasbp_mean | numeric | 3622 | 0.18 % | × |
| meanbp_min | numeric | 103 | 0.00 % | × |
| meanbp_max | numeric | 197 | 0.00 % | × |
| meanbp_mean | numeric | 3677 | 0.00 % | × |
| resprate_min | numeric | 31 | 0.02 % | × |
| resprate_max | numeric | 54 | 0.02 % | × |
| resprate_mean | numeric | 3111 | 0.02 % | × |
| tempc_min | numeric | 168 | 2.26 % | × |
| tempc_max | numeric | 165 | 2.26 % | × |
| tempc_mean | numeric | 1693 | 2.26 % | × |
| spo2_min | numeric | 72 | 0.02 % | × |
| spo2_max | numeric | 13 | 0.02 % | × |
| spo2_mean | numeric | 2016 | 0.02 % | × |
| glucose_min1 | numeric | 246 | 0.66 % | × |
| glucose_max1 | numeric | 455 | 0.66 % | × |
| glucose_mean | numeric | 2229 | 0.66 % | × |
| rrt | numeric | 2 | 0.00 % | |
| subject_id | numeric | 4559 | 0.00 % | × |
| hadm_id…102 | numeric | 4559 | 0.00 % | |
| icustay_id…103 | numeric | 4559 | 0.00 % | |
| urineoutput | numeric | 1794 | 0.00 % | × |
| colloid_bolus | numeric | 14 | 88.84 % | × |
| crystalloid_bolus | numeric | 97 | 26.19 % | × |
Variable list
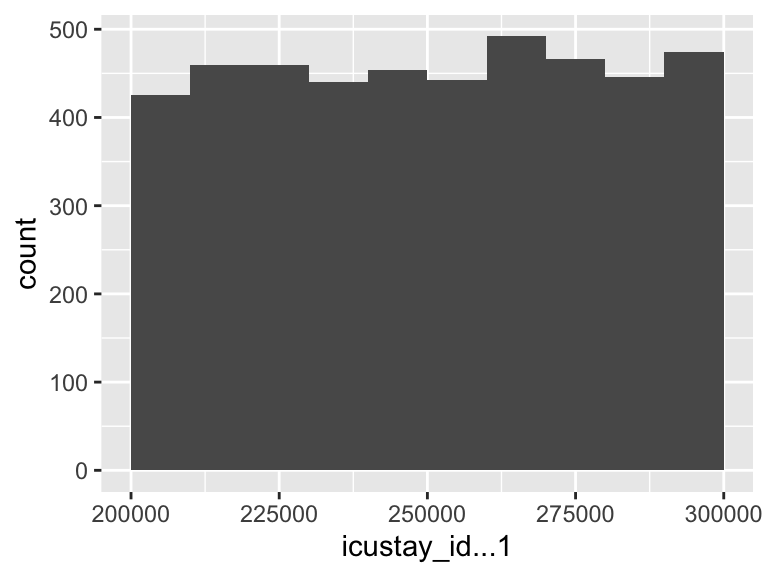
icustay_id…1
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 4559 |
| Median | 251008 |
| 1st and 3rd quartiles | 225575.5; 275526 |
| Min. and max. | 200075; 299998 |
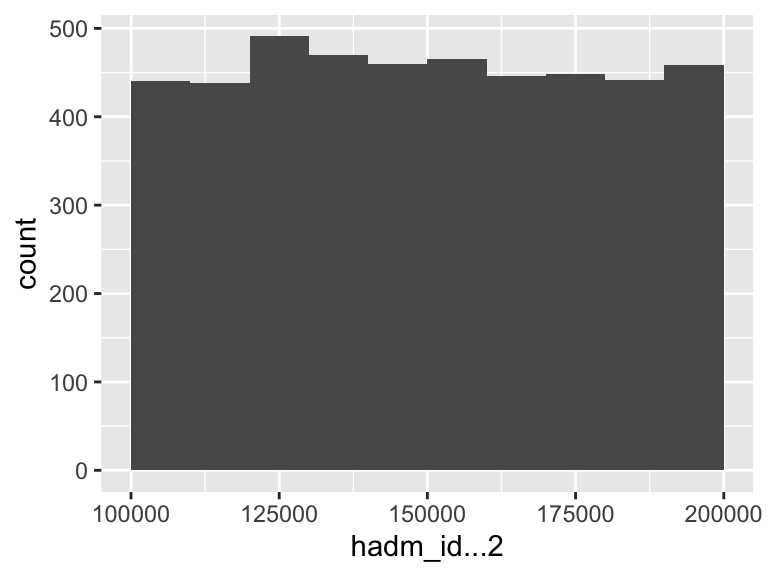
hadm_id…2
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 4559 |
| Median | 149643 |
| 1st and 3rd quartiles | 125389; 175033 |
| Min. and max. | 100003; 199962 |
intime
- The variable is a key (distinct values for each observation).
outtime
- The variable is a key (distinct values for each observation).
dbsource
- The variable only takes one (non-missing) value: "metavision". The variable contains 0 % missing observations.
suspected_infection_time_poe
| Feature | Result |
|---|---|
| Variable type | character |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 4558 |
| Mode | “18/8/2159 00:00:00” |
- Note that the following levels have at most five observations: "1/1/2118 1:02", "1/1/2118 14:36", "1/1/2125 14:05", "1/1/2132 21:00", "1/1/2135 15:55", …, "9/9/2184 10:25", "9/9/2185 18:24", "9/9/2186 18:18", "9/9/2188 1:05", "9/9/2198 18:30" (4548 values omitted).
suspected_infection_time_poe_days
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 4370 |
| Median | 0.03 |
| 1st and 3rd quartiles | -0.08; 0.16 |
| Min. and max. | -0.99; 1 |
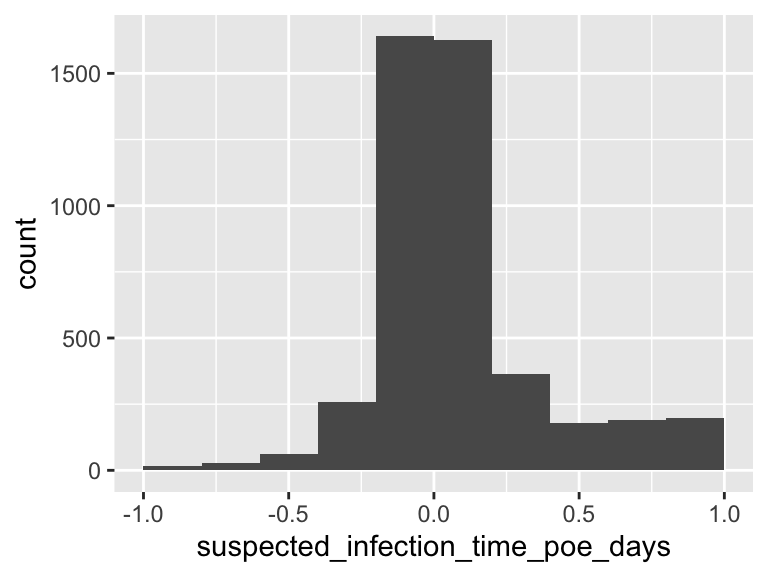
- Note that the following possible outlier values were detected: "-0.99", "-0.98", "-0.93", "-0.93", "-0.92", …, "0.99", "0.99", "0.99", "0.99", "1" (519 values omitted).
specimen_poe
| Feature | Result |
|---|---|
| Variable type | character |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 35 |
| Mode | “BLOOD CULTURE” |
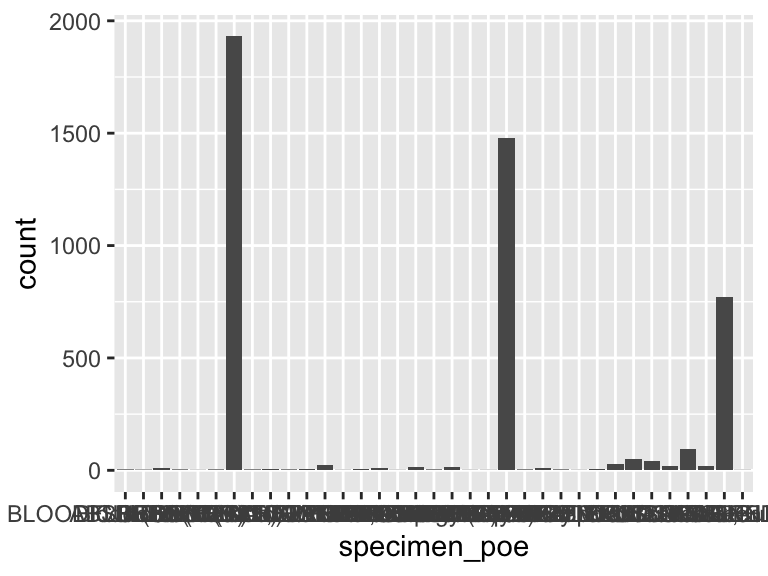
- Note that the following levels have at most five observations: "ABSCESS", "BILE", "Blood (EBV)", "Blood (Malaria)", "Blood (Toxo)", …, "NEOPLASTIC BLOOD", "PLEURAL FLUID", "RAPID RESPIRATORY VIRAL ANTIGEN TEST", "Rapid Respiratory Viral Screen & Culture", "URINE,KIDNEY" (10 values omitted).
positiveculture_poe
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
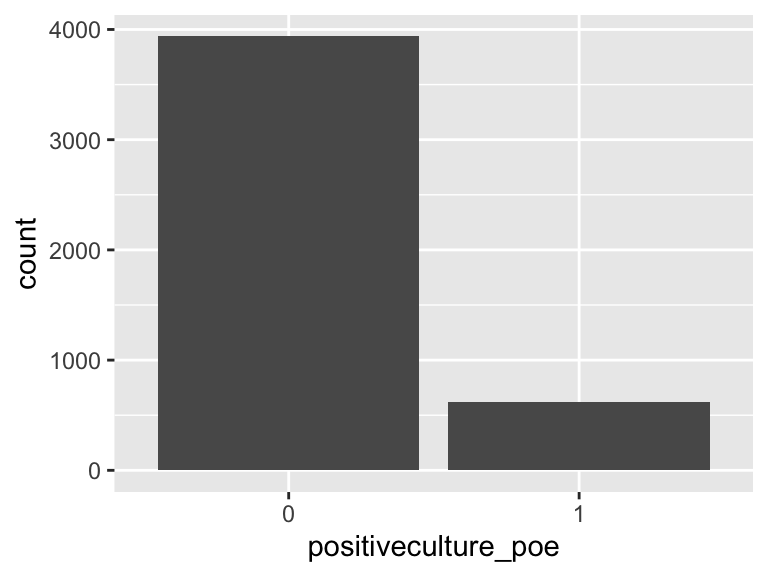
antibiotic_time_poe
| Feature | Result |
|---|---|
| Variable type | character |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 4274 |
| Mode | “1/4/2191 0:00” |
- Note that the following levels have at most five observations: "1/1/2111 0:00", "1/1/2116 0:00", "1/1/2130 0:00", "1/1/2134 0:00", "1/1/2135 0:00", …, "9/9/2181 0:00", "9/9/2185 0:00", "9/9/2187 0:00", "9/9/2189 0:00", "9/9/2198 0:00" (4264 values omitted).
blood_culture_time
| Feature | Result |
|---|---|
| Variable type | character |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 4558 |
| Mode | “18/8/2159 00:00:00” |
- Note that the following levels have at most five observations: "1/1/2113 20:45", "1/1/2118 1:02", "1/1/2118 14:36", "1/1/2125 14:05", "1/1/2132 21:00", …, "9/9/2184 10:25", "9/9/2185 18:24", "9/9/2186 18:18", "9/9/2188 1:05", "9/9/2198 18:30" (4548 values omitted).
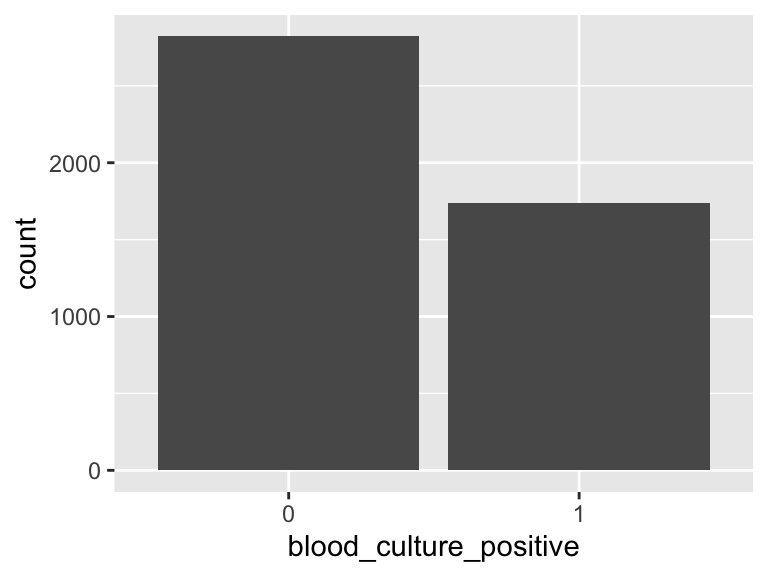
blood_culture_positive
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
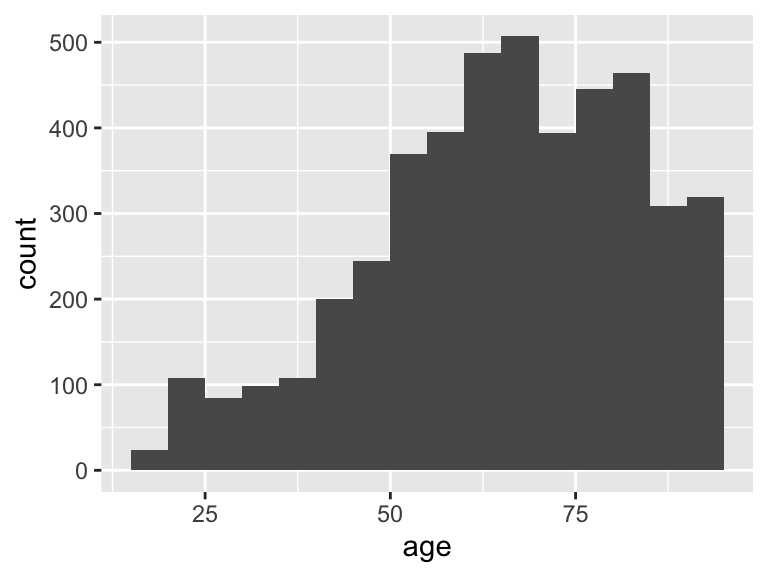
age
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 3831 |
| Median | 66.59 |
| 1st and 3rd quartiles | 53.76; 79.53 |
| Min. and max. | 16.78; 91.4 |
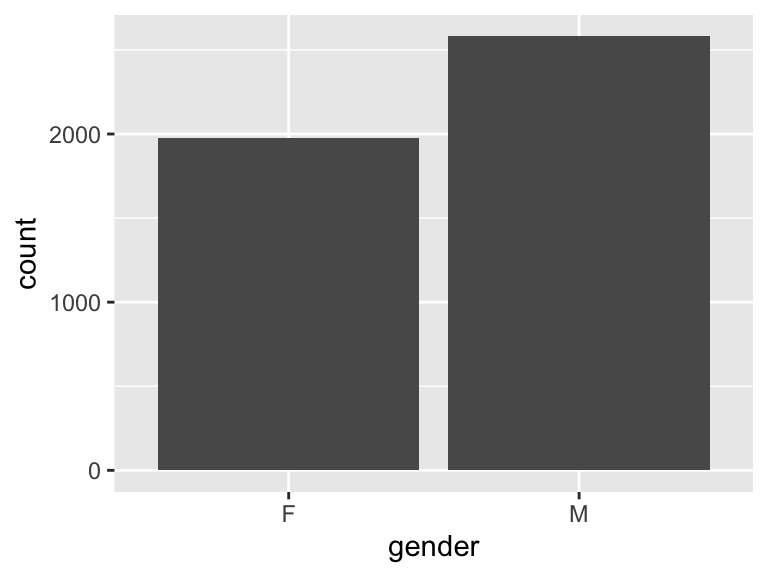
gender
| Feature | Result |
|---|---|
| Variable type | character |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “M” |
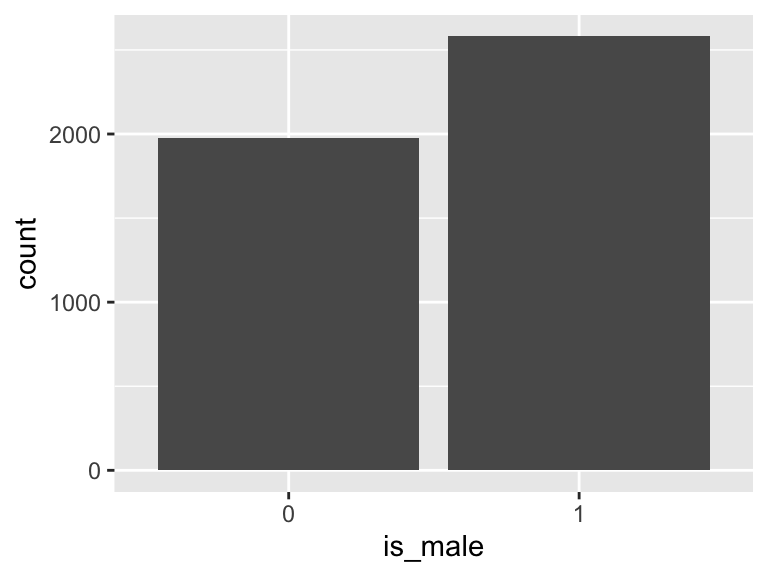
is_male
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “1” |
| Reference category | 0 |
ethnicity
| Feature | Result |
|---|---|
| Variable type | character |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 41 |
| Mode | “WHITE” |
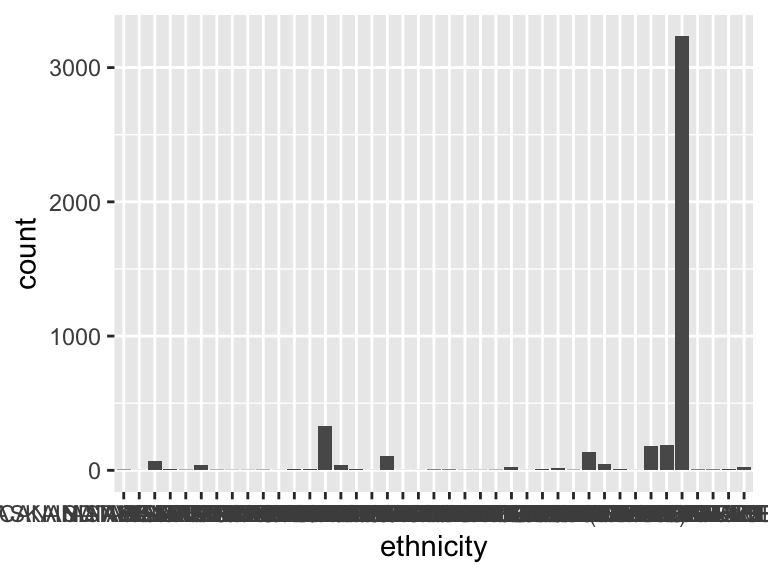
- Note that the following levels have at most five observations: "AMERICAN INDIAN/ALASKA NATIVE", "AMERICAN INDIAN/ALASKA NATIVE FEDERALLY RECOGNIZED TRIBE", "ASIAN - CAMBODIAN", "ASIAN - FILIPINO", "ASIAN - JAPANESE", …, "HISPANIC/LATINO - SALVADORAN", "NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER", "SOUTH AMERICAN", "WHITE - BRAZILIAN", "WHITE - EASTERN EUROPEAN" (11 values omitted).
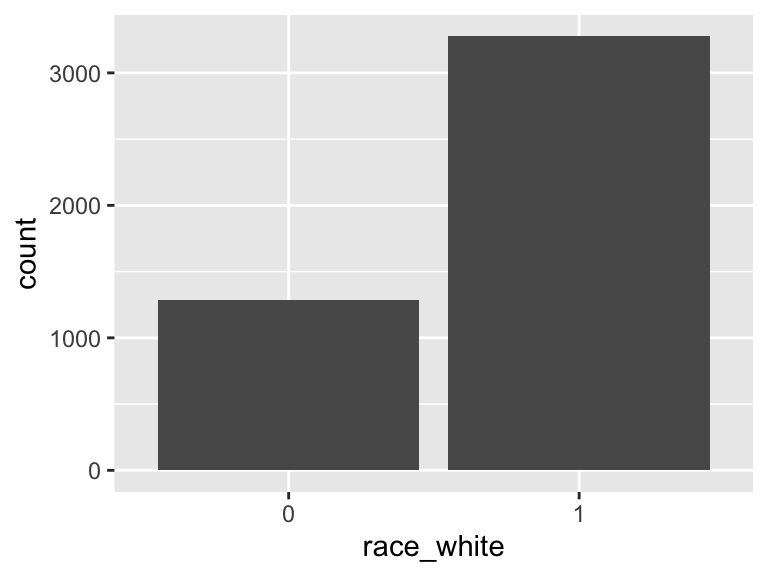
race_white
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “1” |
| Reference category | 0 |
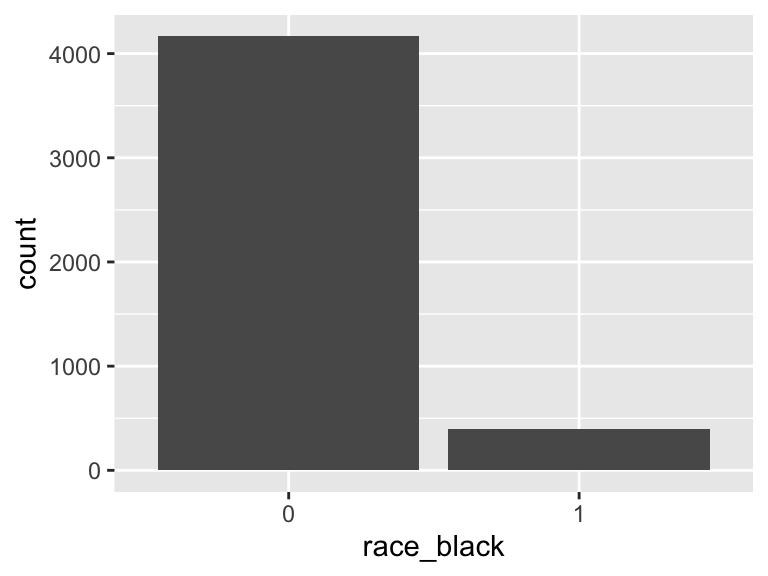
race_black
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
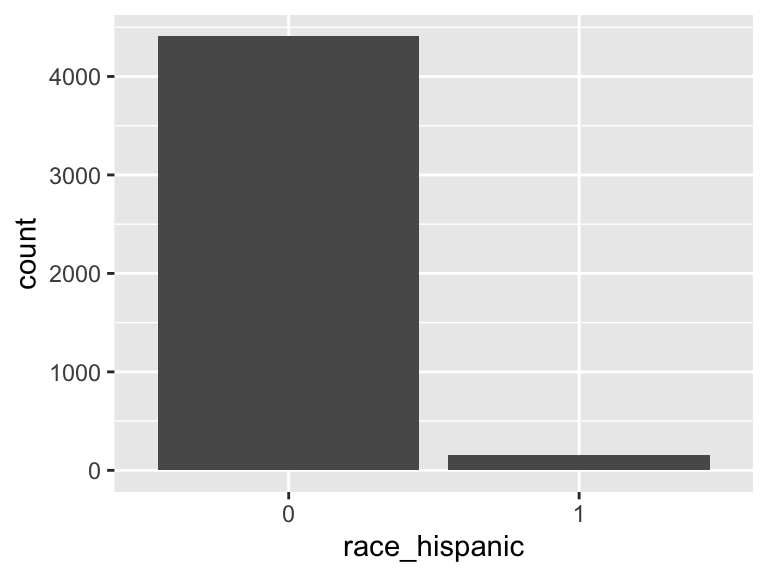
race_hispanic
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
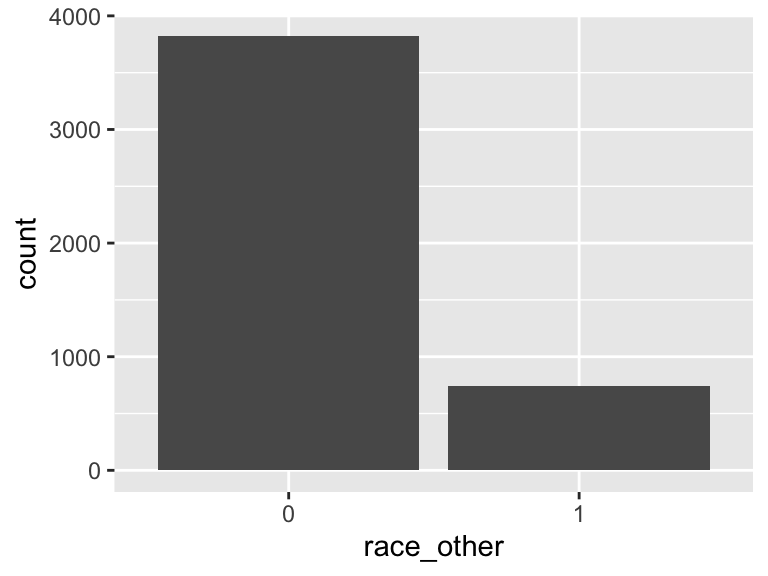
race_other
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
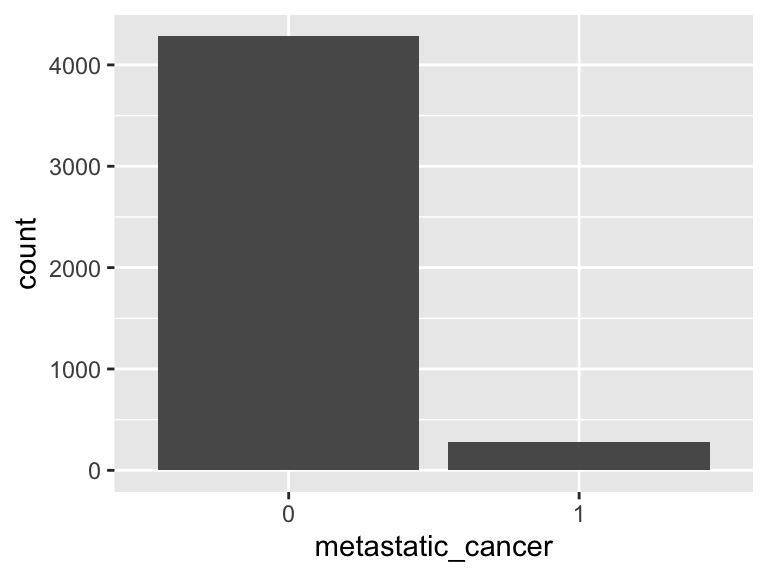
metastatic_cancer
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
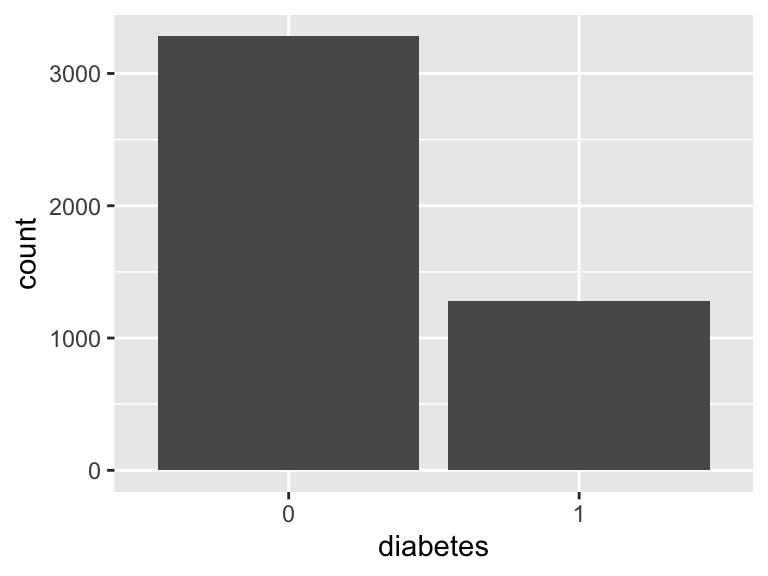
diabetes
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
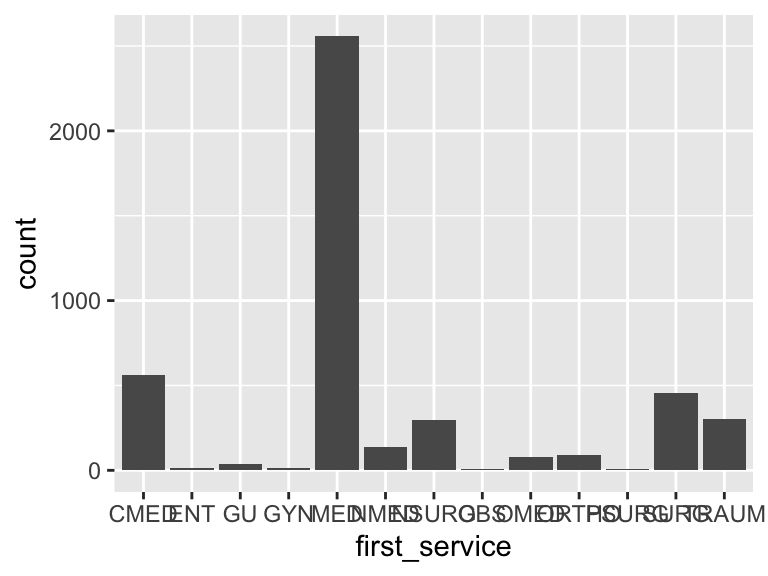
first_service
| Feature | Result |
|---|---|
| Variable type | character |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 13 |
| Mode | “MED” |
hospital_expire_flag
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
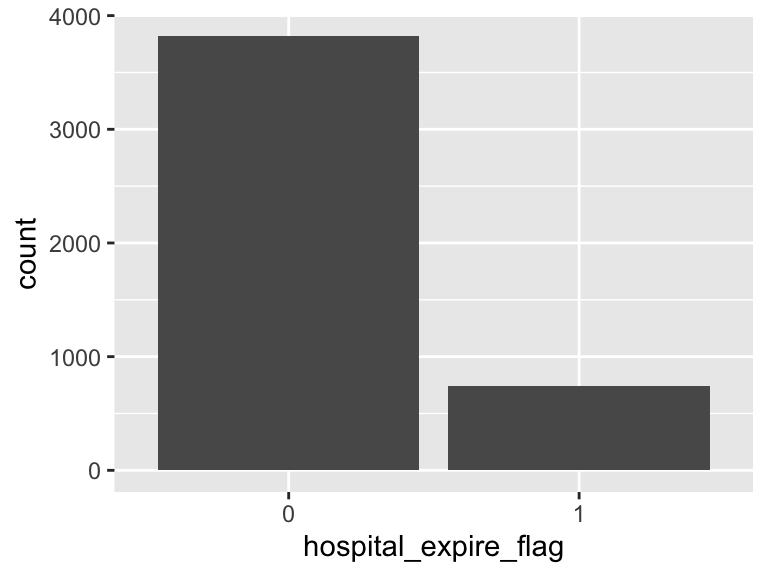
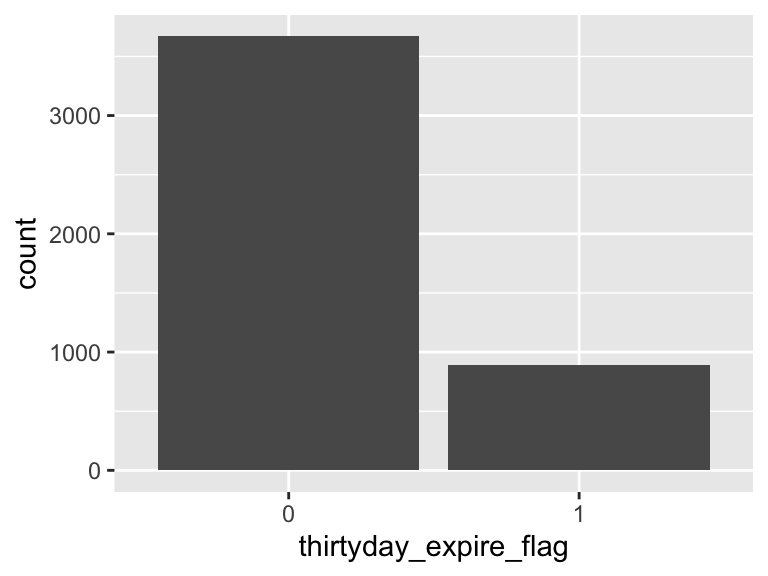
thirtyday_expire_flag
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
icu_los
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 4401 |
| Median | 2.82 |
| 1st and 3rd quartiles | 1.61; 5.87 |
| Min. and max. | 0.17; 101.74 |
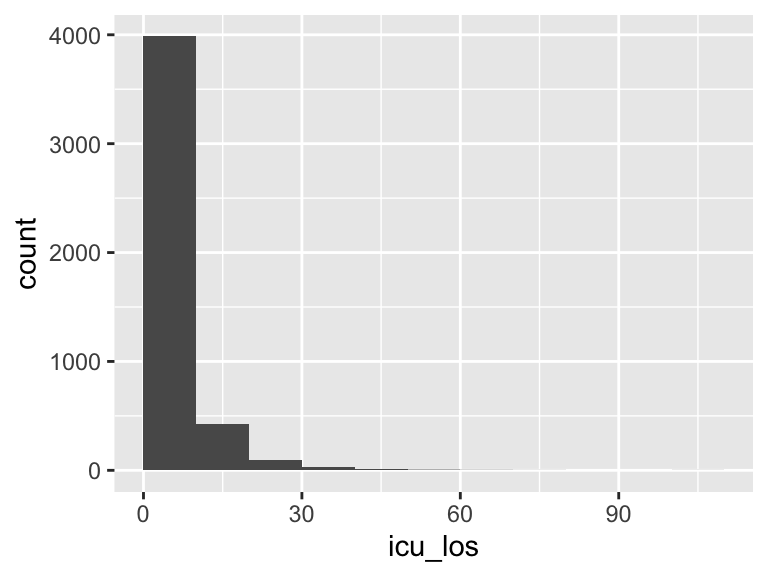
- Note that the following possible outlier values were detected: "0.17", "0.25", "0.25", "0.3", "0.31", …, "59.47", "60.94", "61.93", "79.11", "101.74" (96 values omitted).
hosp_los
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 4191 |
| Median | 7.79 |
| 1st and 3rd quartiles | 4.73; 13.03 |
| Min. and max. | -0.43; 206.43 |
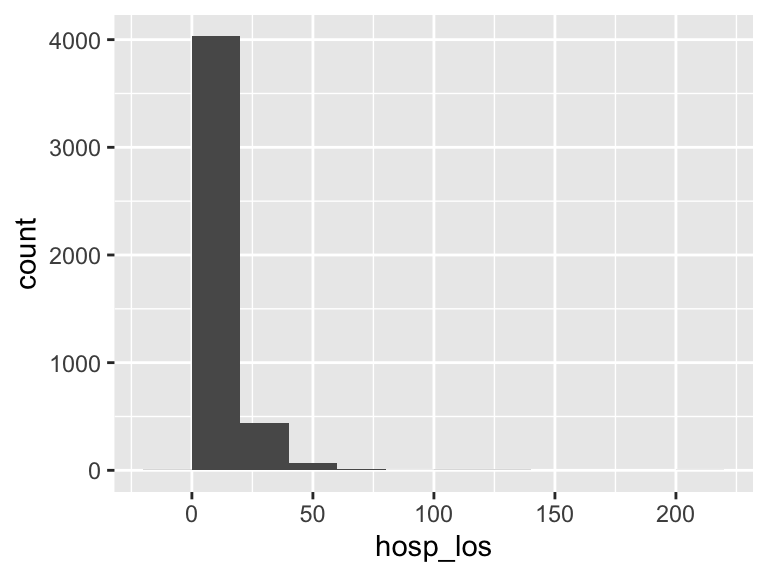
- Note that the following possible outlier values were detected: "-0.43", "-0.06", "0.11", "0.17", "0.18", …, "100.49", "101.62", "123.13", "133.1", "206.43" (207 values omitted).
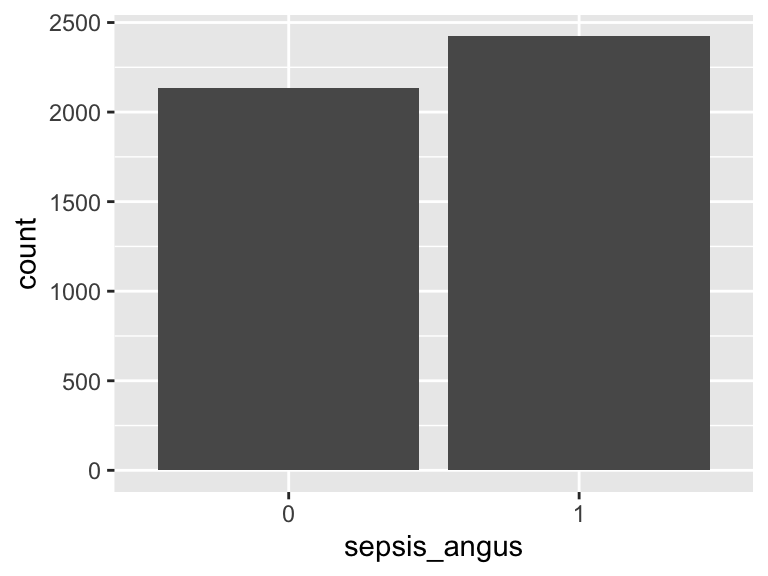
sepsis_angus
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “1” |
| Reference category | 0 |
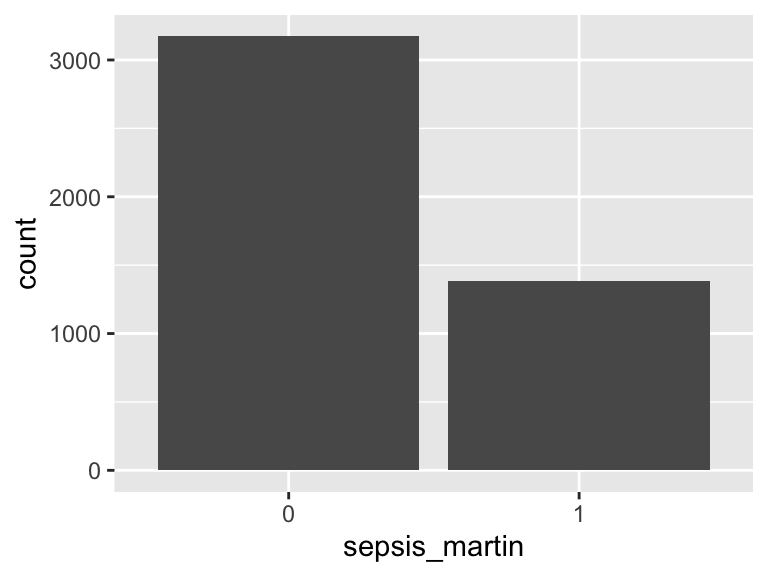
sepsis_martin
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
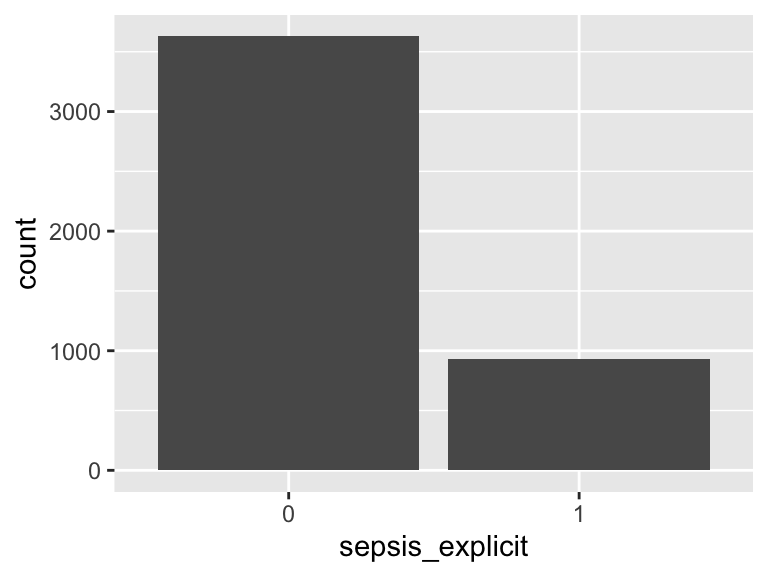
sepsis_explicit
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
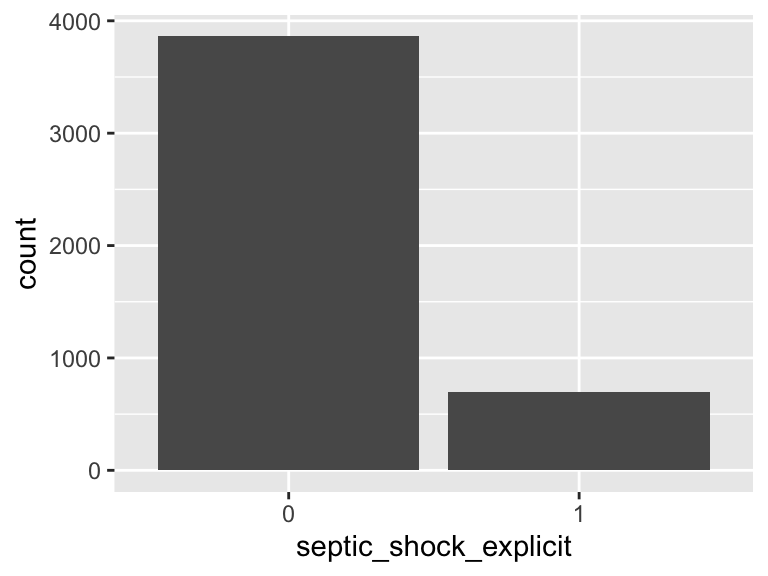
septic_shock_explicit
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
severe_sepsis_explicit
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
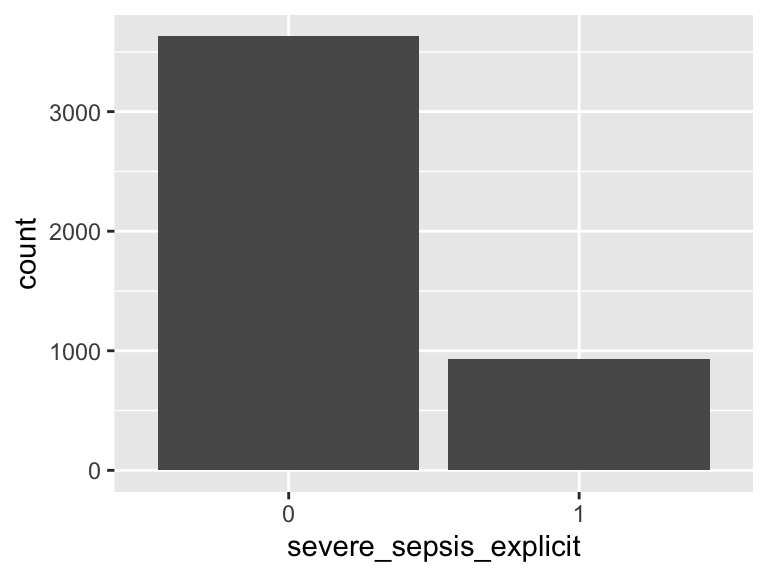
sepsis_nqf
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
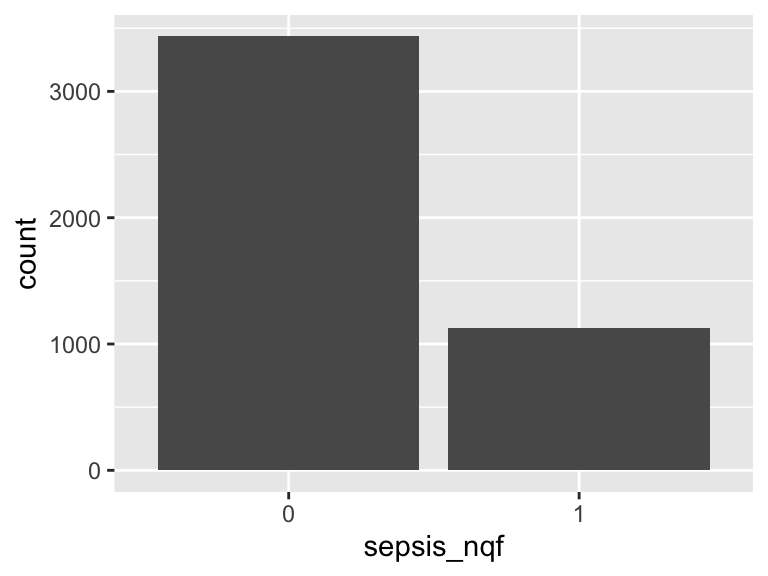
sepsis_cdc
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “1” |
| Reference category | 0 |
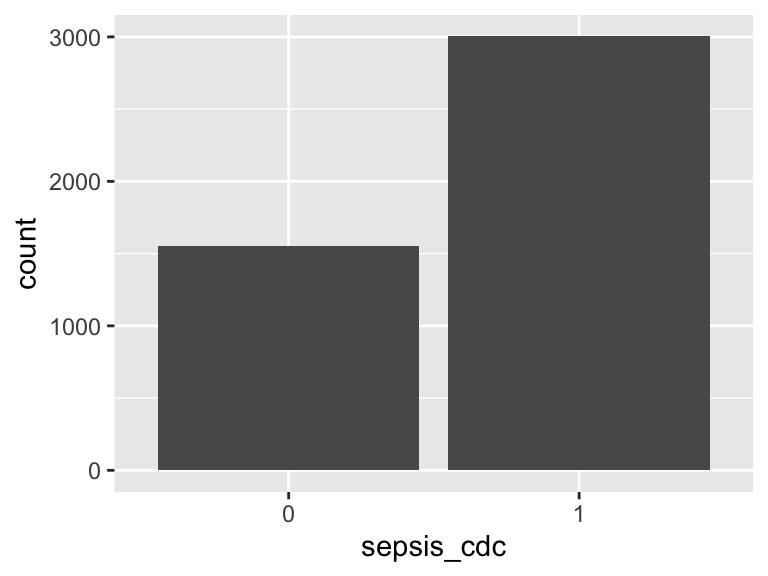
sepsis_cdc_simple
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “1” |
| Reference category | 0 |
elixhauser_hospital
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 49 |
| Median | 4 |
| 1st and 3rd quartiles | 0; 9 |
| Min. and max. | -23; 30 |
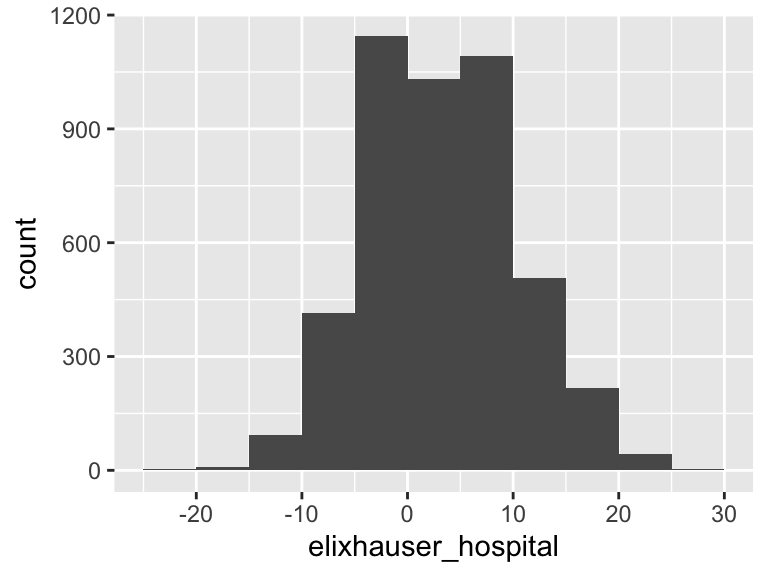
- Note that the following possible outlier values were detected: "-23", "-21", "-20", "-19", "-16", …, "24", "25", "27", "28", "30" (3 values omitted).
vent
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “1” |
| Reference category | 0 |
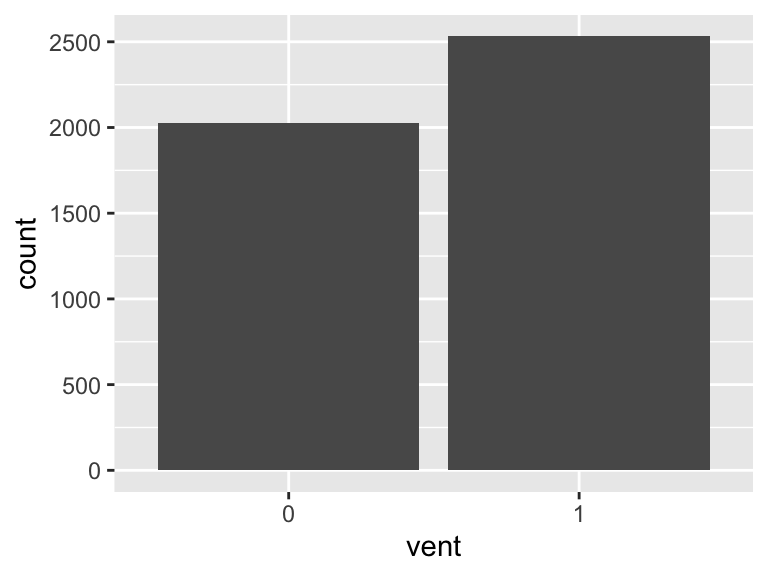
sofa
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 20 |
| Median | 5 |
| 1st and 3rd quartiles | 3; 7 |
| Min. and max. | 2; 21 |
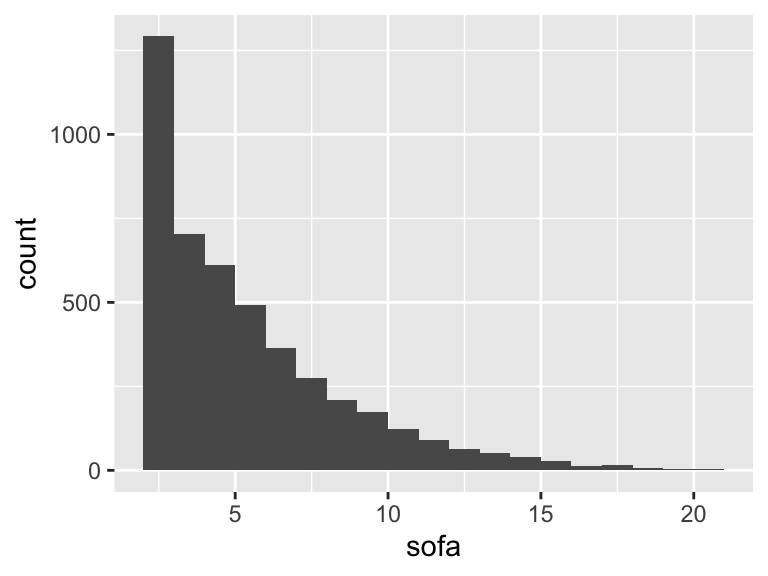
- Note that the following possible outlier values were detected: "18", "19", "20", "21".
lods
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 21 |
| Median | 5 |
| 1st and 3rd quartiles | 3; 7 |
| Min. and max. | 0; 20 |
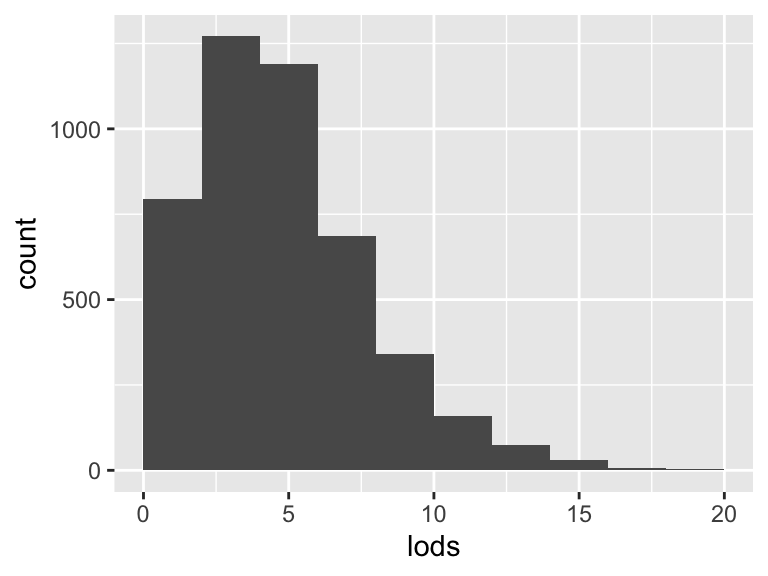
- Note that the following possible outlier values were detected: "14", "15", "16", "17", "18", "19", "20".
sirs
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 5 |
| Mode | “3” |
| Reference category | 0 |
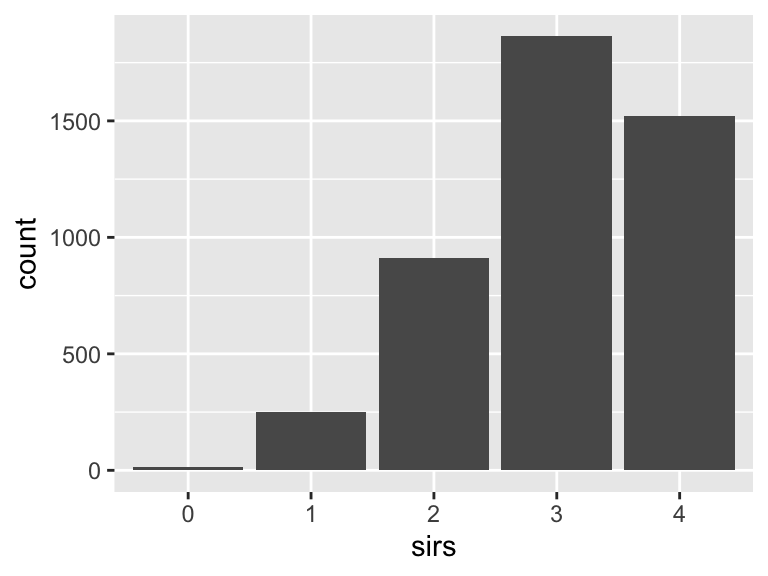
qsofa
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 4 |
| Mode | “2” |
| Reference category | 0 |
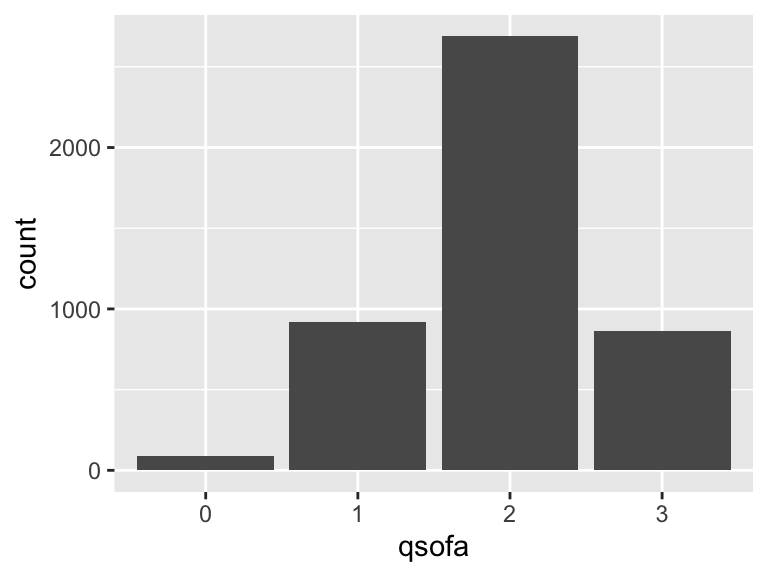
qsofa_sysbp_score
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 7 (0.15 %) |
| Number of unique values | 2 |
| Mode | “1” |
| Reference category | 0 |
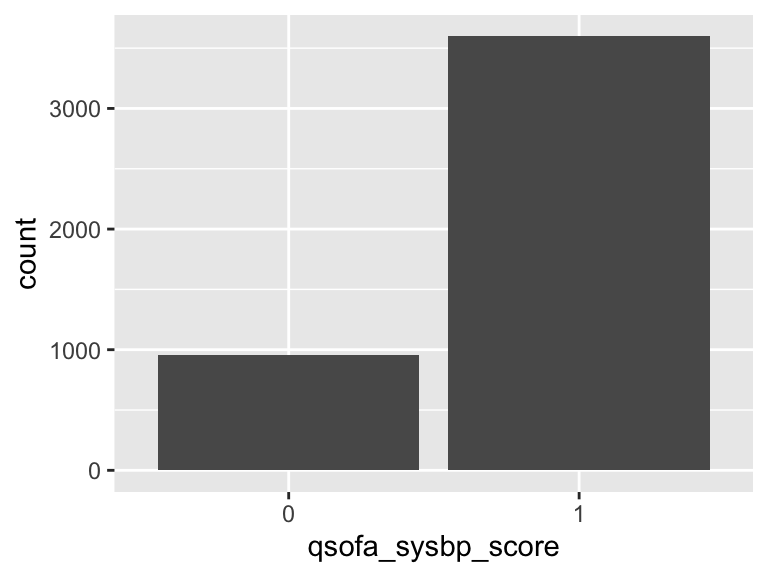
qsofa_gcs_score
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 3 (0.07 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
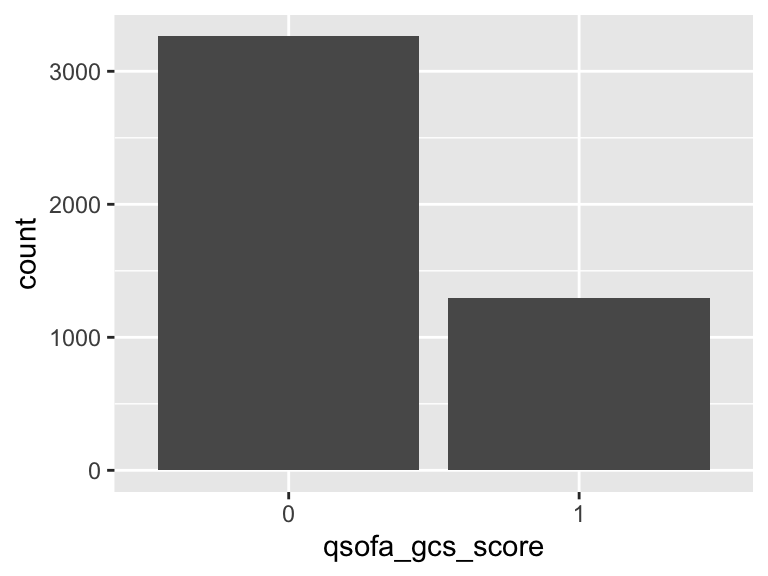
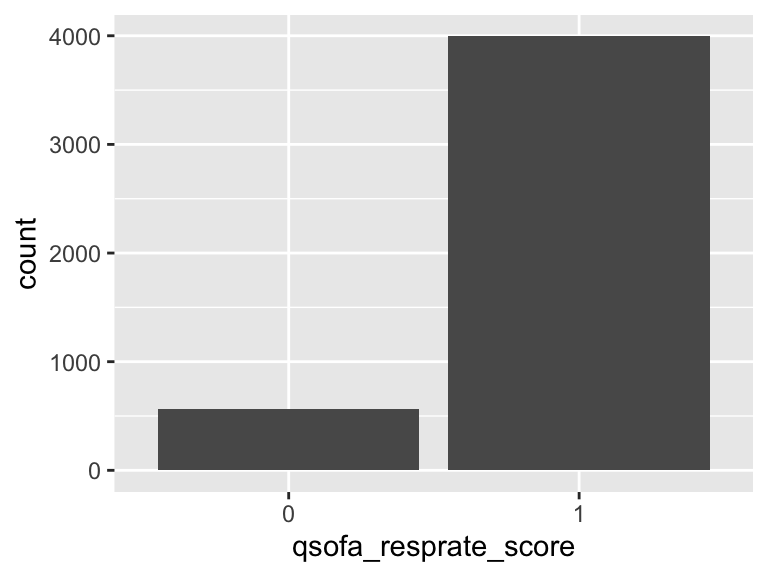
qsofa_resprate_score
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 1 (0.02 %) |
| Number of unique values | 2 |
| Mode | “1” |
| Reference category | 0 |
aniongap_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 14 (0.31 %) |
| Number of unique values | 34 |
| Median | 12 |
| 1st and 3rd quartiles | 10; 15 |
| Min. and max. | 3; 42 |
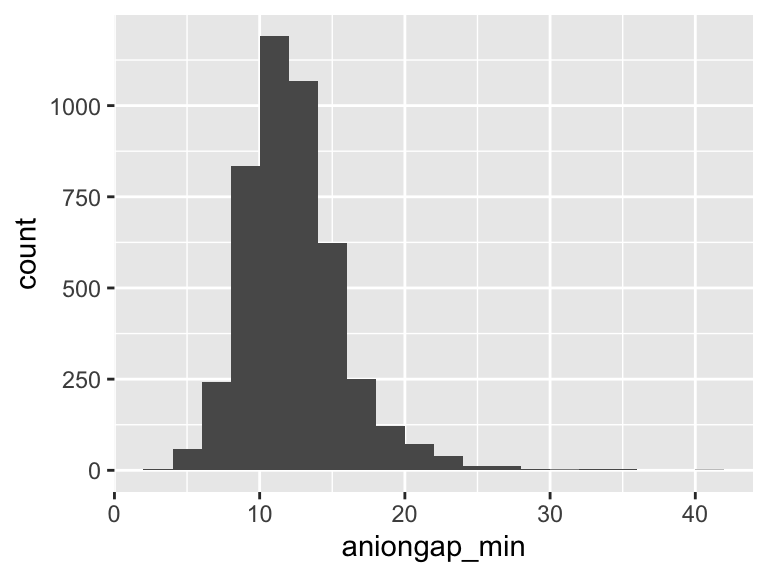
- Note that the following possible outlier values were detected: "3", "4", "5", "6", "29", …, "32", "33", "35", "36", "42" (2 values omitted).
aniongap_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 14 (0.31 %) |
| Number of unique values | 43 |
| Median | 16 |
| 1st and 3rd quartiles | 13; 19 |
| Min. and max. | 6; 56 |
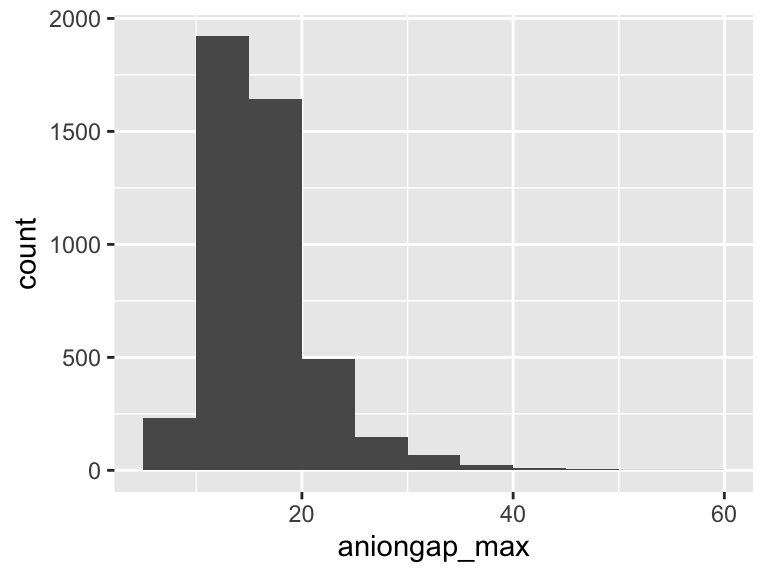
- Note that the following possible outlier values were detected: "6", "7", "32", "33", "34", …, "44", "46", "47", "54", "56" (9 values omitted).
bicarbonate_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 3 (0.07 %) |
| Number of unique values | 39 |
| Median | 21 |
| 1st and 3rd quartiles | 18; 24 |
| Min. and max. | 5; 45 |
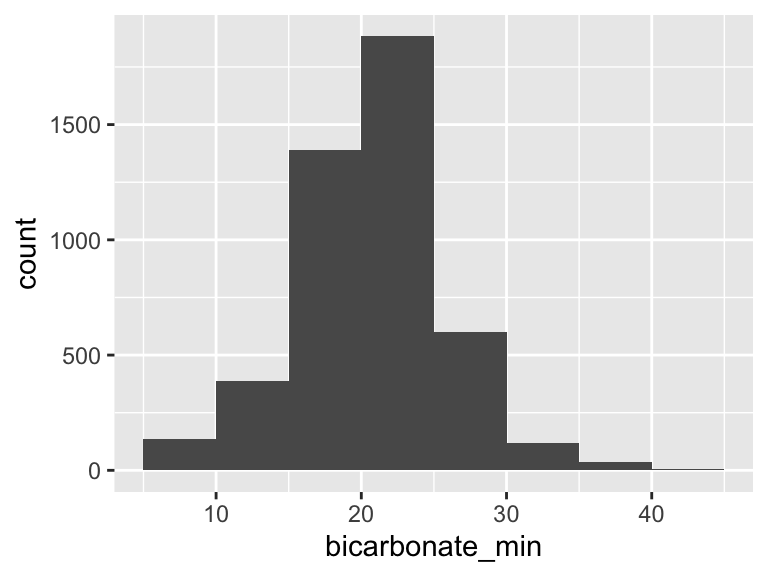
- Note that the following possible outlier values were detected: "5", "6", "7", "8", "34", …, "38", "39", "40", "41", "45" (3 values omitted).
bicarbonate_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 3 (0.07 %) |
| Number of unique values | 42 |
| Median | 24 |
| 1st and 3rd quartiles | 22; 27 |
| Min. and max. | 8; 49 |
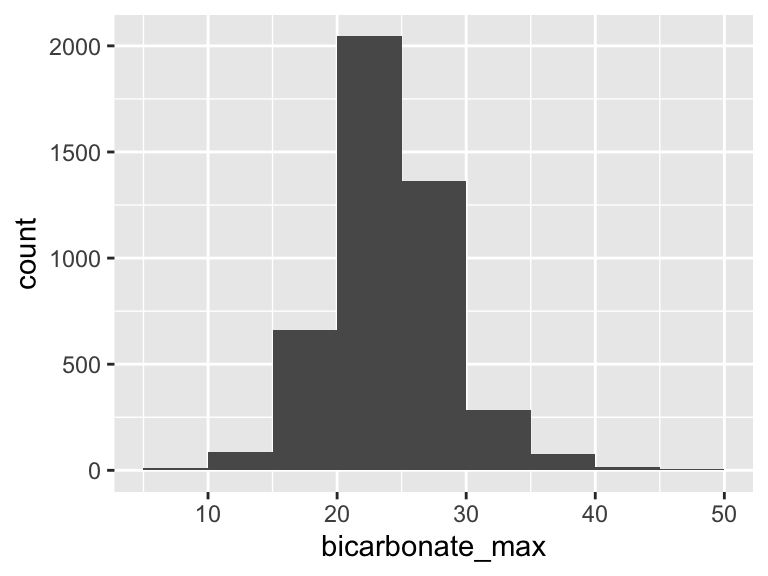
- Note that the following possible outlier values were detected: "8", "9", "10", "11", "12", …, "44", "45", "46", "47", "49" (11 values omitted).
creatinine_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 2 (0.04 %) |
| Number of unique values | 97 |
| Median | 1 |
| 1st and 3rd quartiles | 0.7; 1.4 |
| Min. and max. | 0.1; 22.1 |
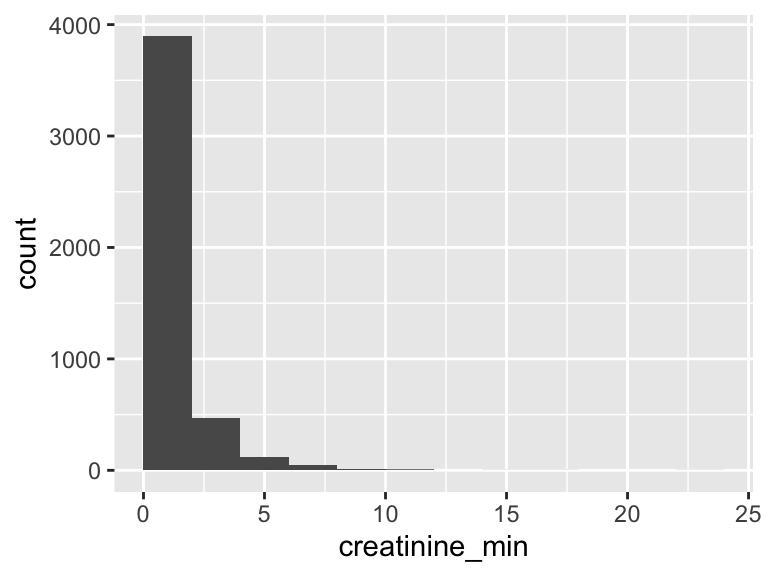
- Note that the following possible outlier values were detected: "0.1", "0.2", "0.3", "0.4", "4.3", …, "11.3", "12", "14.9", "17.3", "22.1" (49 values omitted).
creatinine_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 2 (0.04 %) |
| Number of unique values | 122 |
| Median | 1.2 |
| 1st and 3rd quartiles | 0.9; 1.9 |
| Min. and max. | 0.1; 27.8 |
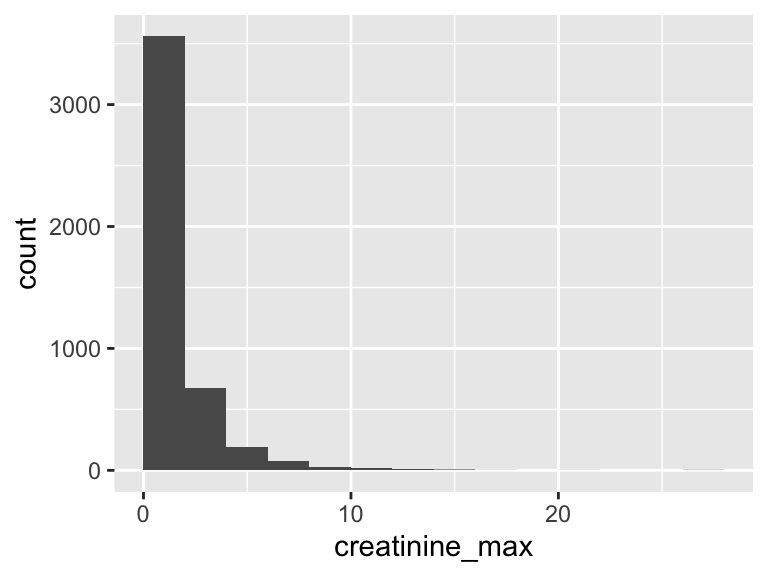
- Note that the following possible outlier values were detected: "0.1", "0.2", "0.3", "0.4", "0.5", …, "16", "17", "20.6", "27.5", "27.8" (45 values omitted).
chloride_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 1 (0.02 %) |
| Number of unique values | 66 |
| Median | 102 |
| 1st and 3rd quartiles | 98; 106 |
| Min. and max. | 58; 139 |
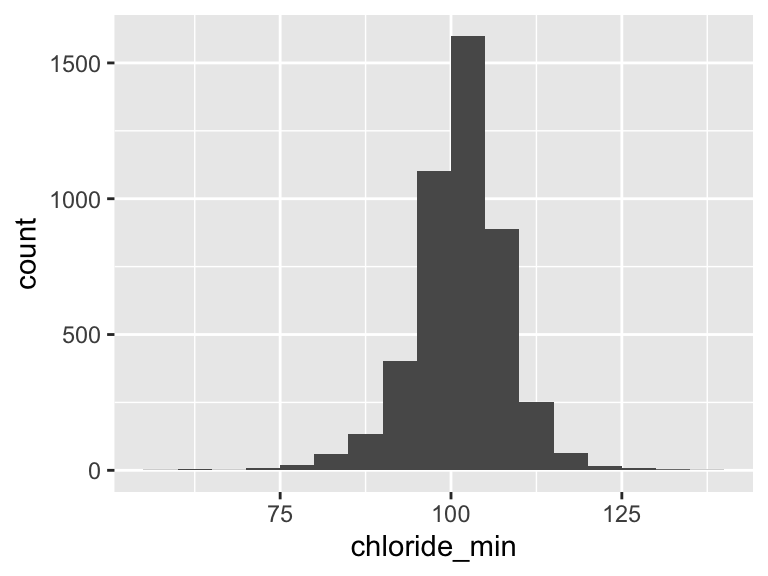
- Note that the following possible outlier values were detected: "58", "61", "62", "64", "65", …, "127", "128", "133", "134", "139" (23 values omitted).
chloride_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 1 (0.02 %) |
| Number of unique values | 64 |
| Median | 108 |
| 1st and 3rd quartiles | 104; 112 |
| Min. and max. | 67; 155 |
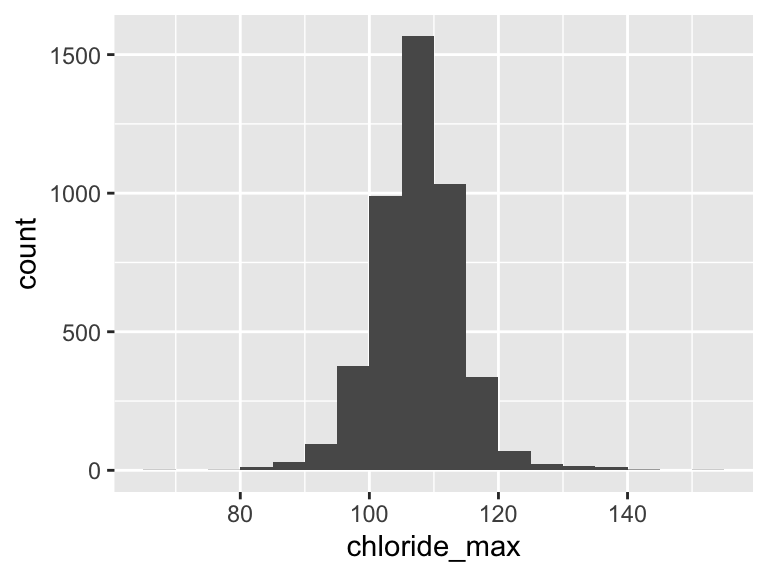
- Note that the following possible outlier values were detected: "67", "80", "81", "82", "84", …, "139", "140", "141", "144", "155" (21 values omitted).
glucose_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 257 |
| Median | 106 |
| 1st and 3rd quartiles | 90; 127 |
| Min. and max. | 15; 425 |
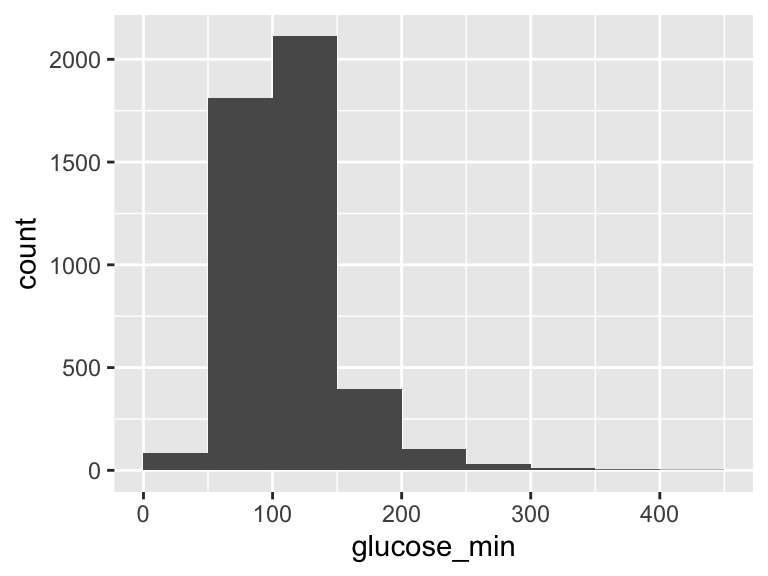
- Note that the following possible outlier values were detected: "15", "19", "20", "21", "22", …, "353", "355", "359", "424", "425" (91 values omitted).
glucose_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 473 |
| Median | 161 |
| 1st and 3rd quartiles | 128; 211 |
| Min. and max. | 38; 2275 |
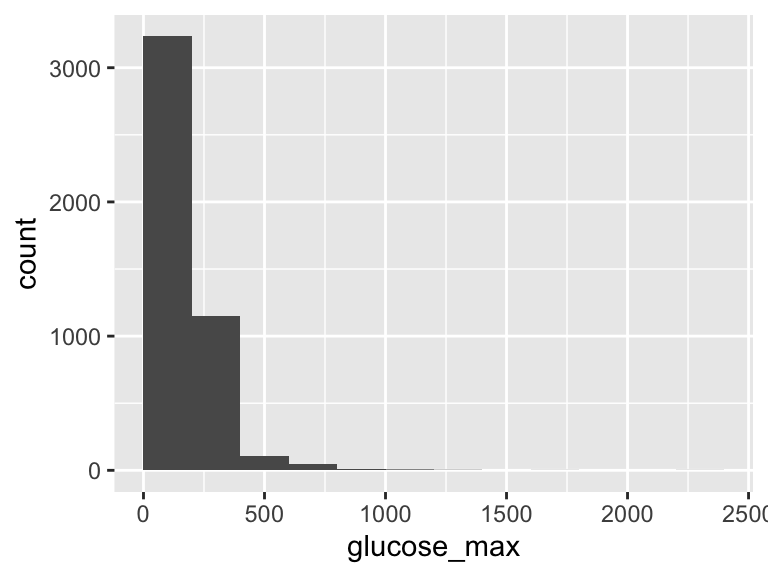
- Note that the following possible outlier values were detected: "38", "40", "56", "57", "58", …, "1155", "1208", "1317", "1689", "2275" (107 values omitted).
hematocrit_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 3 (0.07 %) |
| Number of unique values | 340 |
| Median | 29.5 |
| 1st and 3rd quartiles | 25.3; 34 |
| Min. and max. | 9; 55.9 |
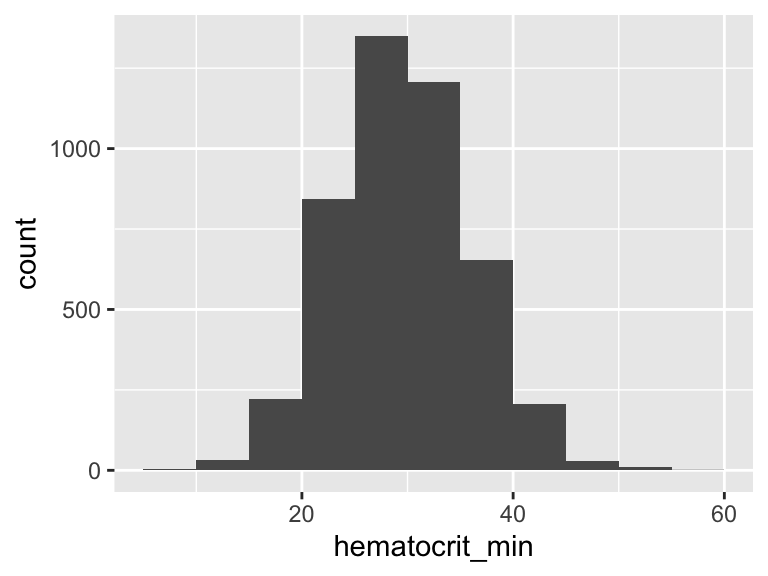
- Note that the following possible outlier values were detected: "9", "9.1", "9.9", "10.6", "11", …, "52.8", "53.3", "54", "54.5", "55.9" (18 values omitted).
hematocrit_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 3 (0.07 %) |
| Number of unique values | 330 |
| Median | 35.6 |
| 1st and 3rd quartiles | 31.6; 40 |
| Min. and max. | 16.4; 61 |
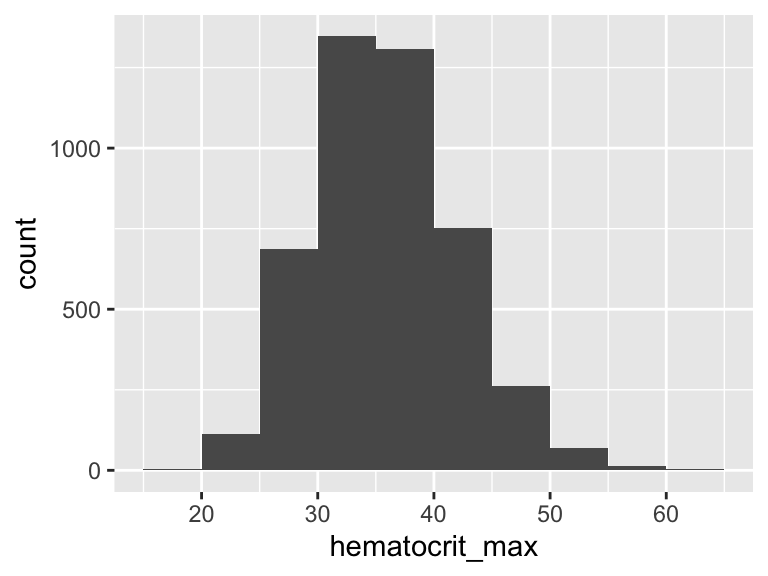
- Note that the following possible outlier values were detected: "16.4", "18.1", "19.1", "20.3", "20.6", …, "59.1", "60.4", "60.5", "60.9", "61" (14 values omitted).
hemoglobin_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 3 (0.07 %) |
| Number of unique values | 138 |
| Median | 9.9 |
| 1st and 3rd quartiles | 8.5; 11.4 |
| Min. and max. | 2.7; 18 |
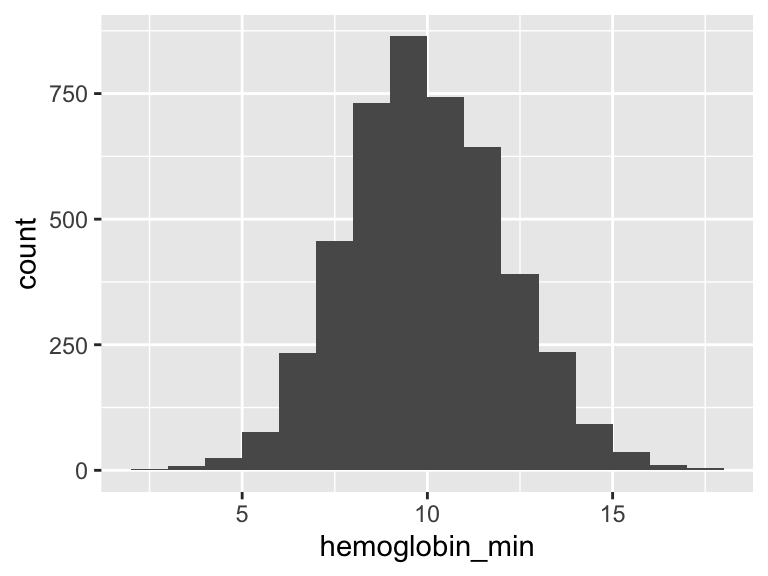
- Note that the following possible outlier values were detected: "2.7", "2.9", "3", "3.2", "3.5", …, "16.6", "17.2", "17.4", "17.5", "18" (14 values omitted).
hemoglobin_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 3 (0.07 %) |
| Number of unique values | 129 |
| Median | 11.85 |
| 1st and 3rd quartiles | 10.4; 13.4 |
| Min. and max. | 4.8; 19.6 |
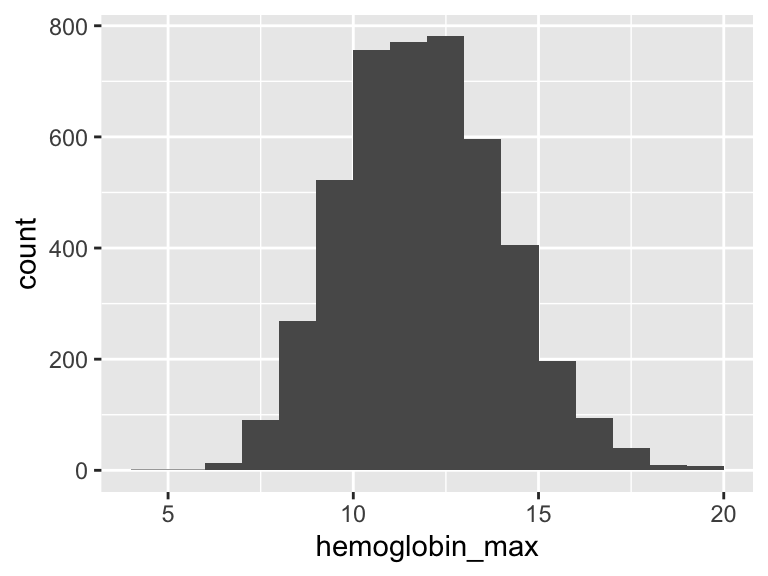
- Note that the following possible outlier values were detected: "4.8", "6", "6.3", "6.4", "18.5", …, "19.1", "19.3", "19.4", "19.5", "19.6" (1 values omitted).
lactate_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 97 |
| Median | 1.4 |
| 1st and 3rd quartiles | 1; 2 |
| Min. and max. | 0.3; 20.3 |
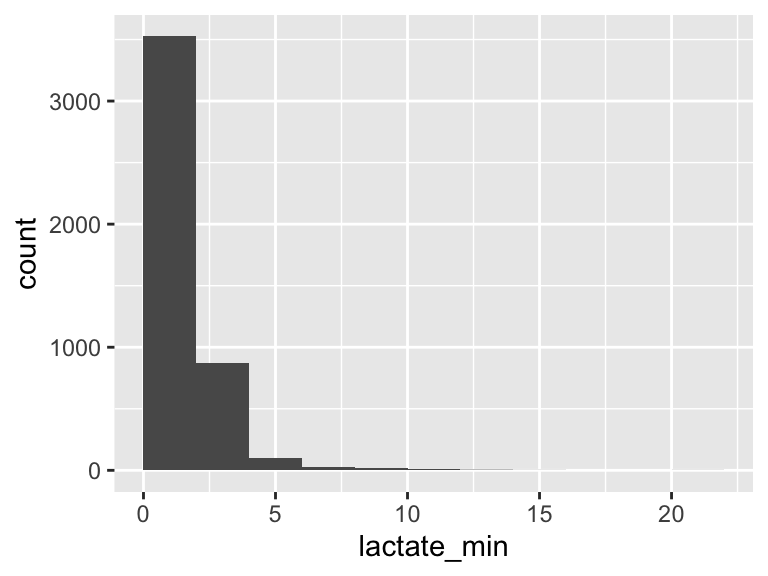
- Note that the following possible outlier values were detected: "0.3", "0.4", "5.4", "5.5", "5.6", …, "12.1", "12.6", "13.2", "15.6", "20.3" (37 values omitted).
lactate_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 165 |
| Median | 2.2 |
| 1st and 3rd quartiles | 1.5; 3.5 |
| Min. and max. | 0.3; 32 |
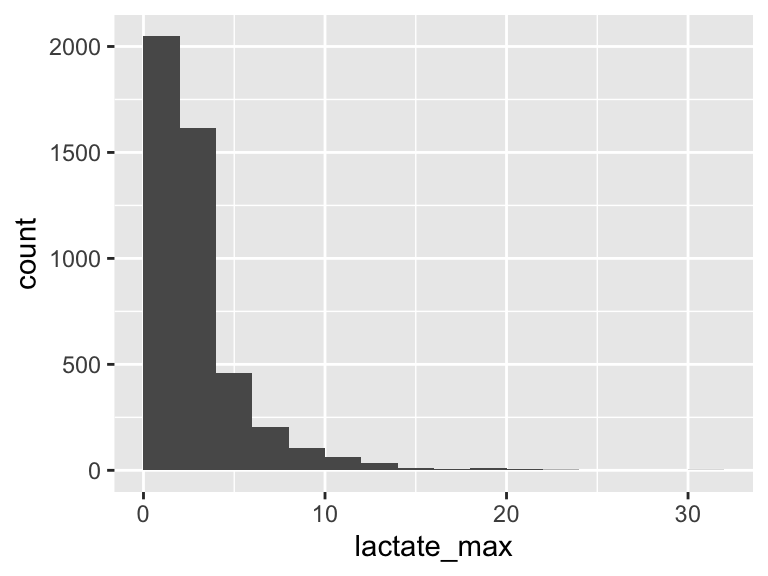
- Note that the following possible outlier values were detected: "0.3", "0.4", "0.5", "0.6", "0.7", …, "21.2", "21.4", "22.4", "23.5", "32" (40 values omitted).
lactate_mean
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 209 |
| Median | 1.9 |
| 1st and 3rd quartiles | 1.35; 2.75 |
| Min. and max. | 0.3; 20.85 |
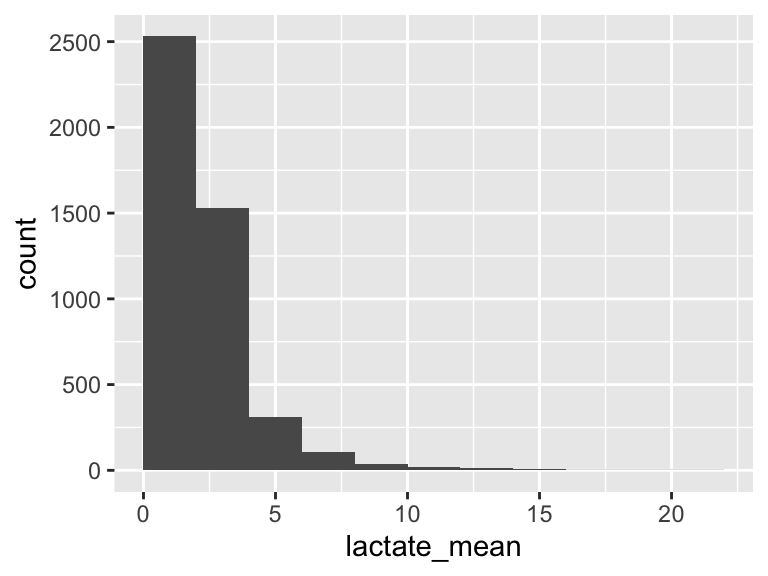
- Note that the following possible outlier values were detected: "0.3", "0.4", "0.45", "0.5", "0.55", …, "15.35", "15.55", "16.8", "19.55", "20.85" (58 values omitted).
platelet_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 6 (0.13 %) |
| Number of unique values | 527 |
| Median | 178 |
| 1st and 3rd quartiles | 122; 246 |
| Min. and max. | 5; 1297 |
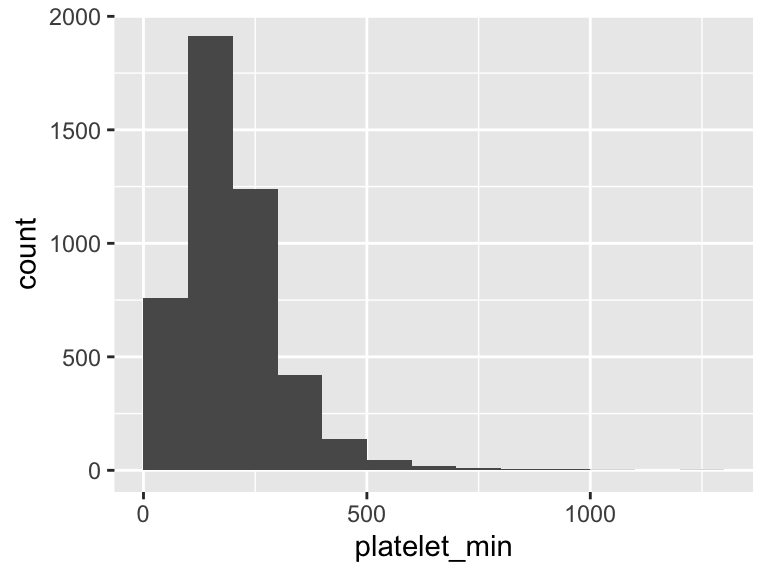
- Note that the following possible outlier values were detected: "5", "6", "7", "8", "9", …, "951", "961", "996", "1039", "1297" (57 values omitted).
platelet_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 6 (0.13 %) |
| Number of unique values | 608 |
| Median | 224 |
| 1st and 3rd quartiles | 159; 303 |
| Min. and max. | 16; 1775 |
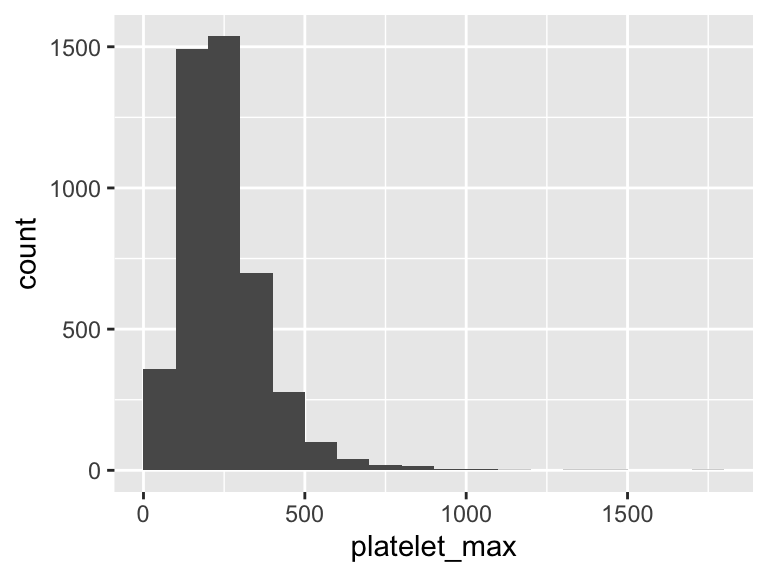
- Note that the following possible outlier values were detected: "16", "18", "19", "20", "21", …, "1081", "1163", "1308", "1418", "1775" (73 values omitted).
potassium_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 51 |
| Median | 3.7 |
| 1st and 3rd quartiles | 3.4; 4.1 |
| Min. and max. | 0.9; 6.8 |
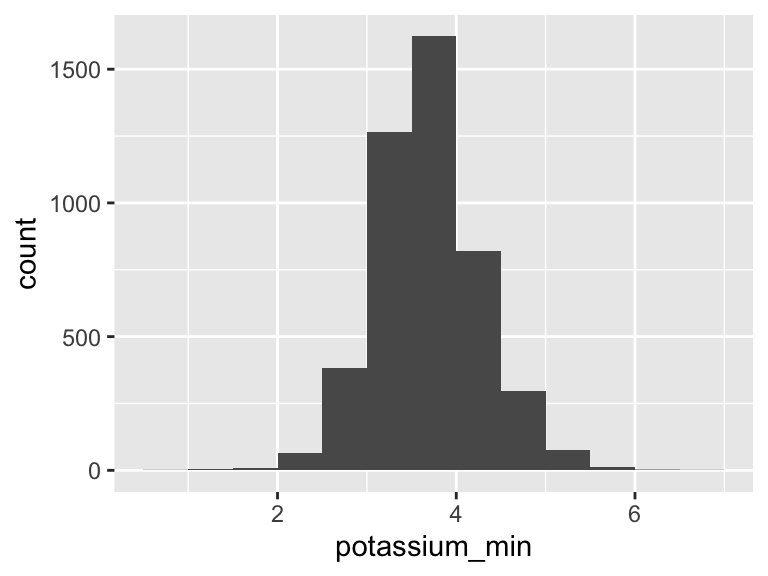
- Note that the following possible outlier values were detected: "0.9", "1.2", "1.4", "1.5", "1.7", …, "5.9", "6", "6.2", "6.4", "6.8" (13 values omitted).
potassium_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 73 |
| Median | 4.5 |
| 1st and 3rd quartiles | 4.1; 5.1 |
| Min. and max. | 2.3; 13.5 |
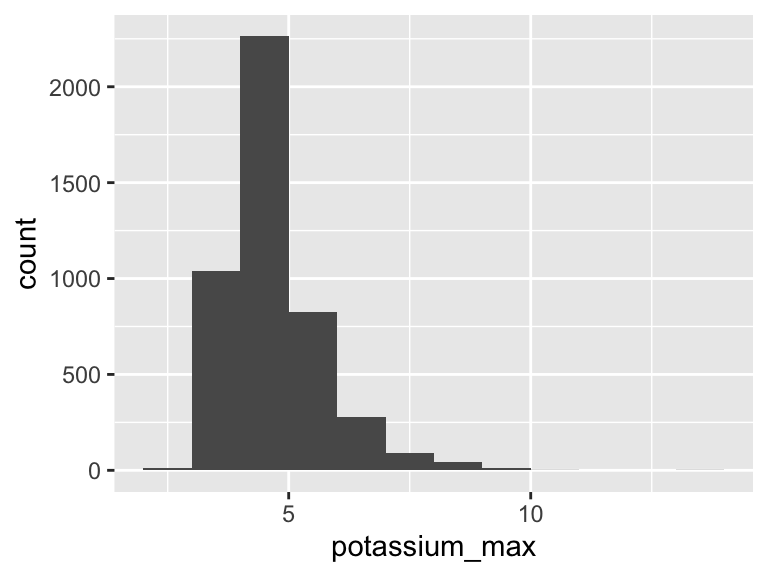
- Note that the following possible outlier values were detected: "2.3", "2.8", "2.9", "3", "3.1", …, "9.7", "9.8", "9.9", "10.2", "13.5" (17 values omitted).
inr_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 270 (5.92 %) |
| Number of unique values | 68 |
| Median | 1.2 |
| 1st and 3rd quartiles | 1.1; 1.4 |
| Min. and max. | 0.6; 14.6 |
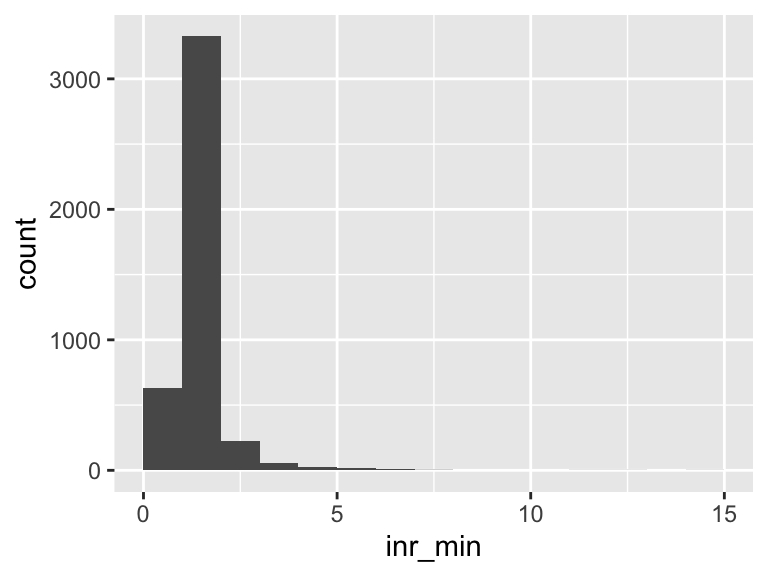
- Note that the following possible outlier values were detected: "0.6", "0.7", "0.8", "0.9", "2.7", …, "7.1", "7.7", "12", "13", "14.6" (38 values omitted).
inr_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 270 (5.92 %) |
| Number of unique values | 98 |
| Median | 1.3 |
| 1st and 3rd quartiles | 1.2; 1.7 |
| Min. and max. | 0.6; 24 |
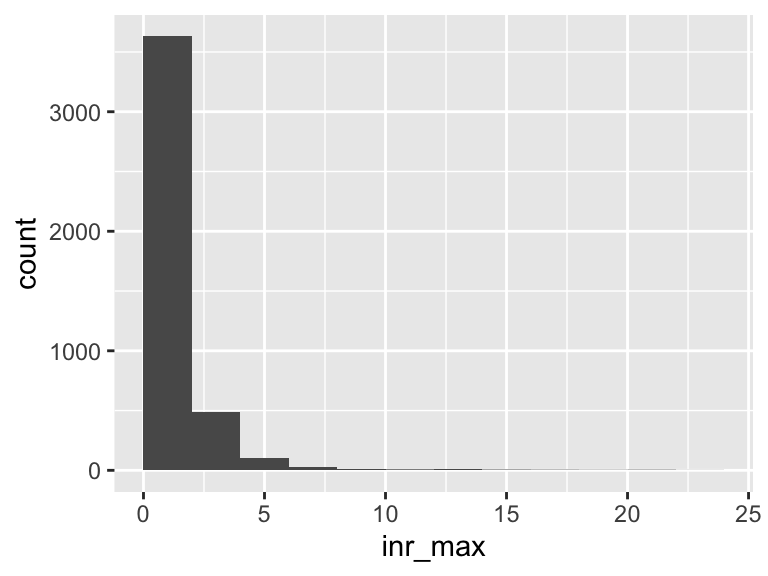
- Note that the following possible outlier values were detected: "0.6", "0.8", "0.9", "1", "5.1", …, "18.8", "21", "21.1", "21.5", "24" (47 values omitted).
sodium_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 61 |
| Median | 136 |
| 1st and 3rd quartiles | 133; 139 |
| Min. and max. | 74; 166 |
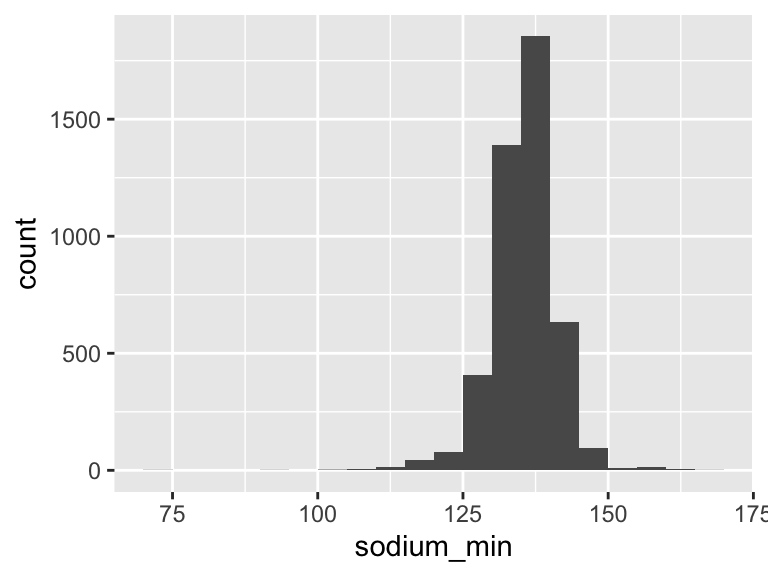
- Note that the following possible outlier values were detected: "74", "95", "103", "104", "106", …, "160", "162", "163", "165", "166" (26 values omitted).
sodium_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 63 |
| Median | 140 |
| 1st and 3rd quartiles | 138; 143 |
| Min. and max. | 108; 182 |
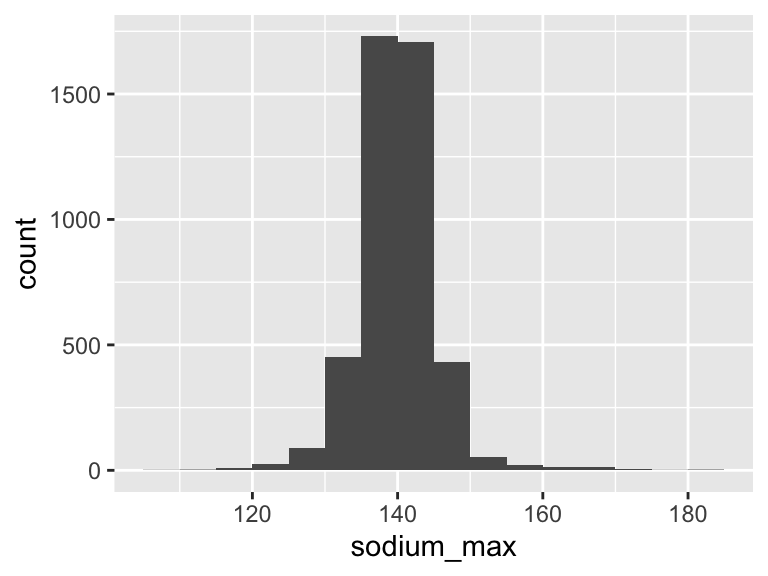
- Note that the following possible outlier values were detected: "108", "113", "114", "116", "117", …, "173", "174", "175", "180", "182" (33 values omitted).
bun_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 2 (0.04 %) |
| Number of unique values | 139 |
| Median | 19 |
| 1st and 3rd quartiles | 13; 32 |
| Min. and max. | 1; 182 |
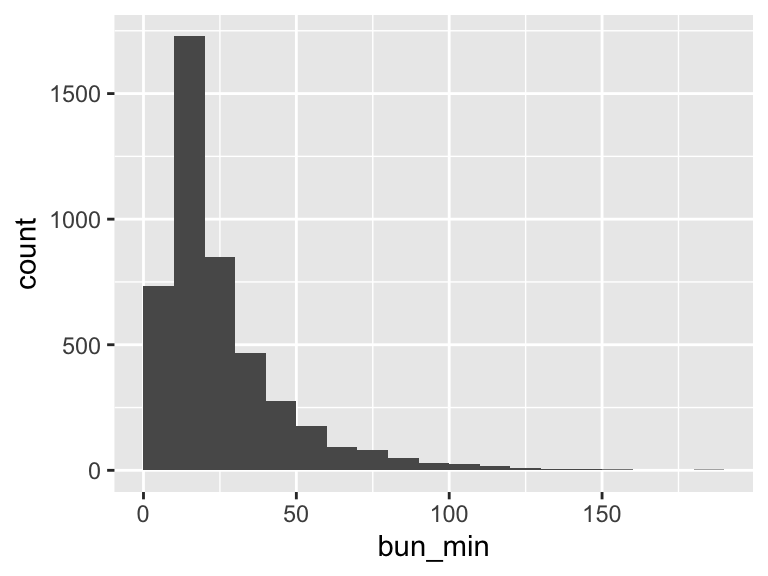
- Note that the following possible outlier values were detected: "1", "2", "3", "4", "5", …, "152", "155", "160", "181", "182" (16 values omitted).
bun_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 2 (0.04 %) |
| Number of unique values | 156 |
| Median | 24 |
| 1st and 3rd quartiles | 16; 39 |
| Min. and max. | 3; 261 |
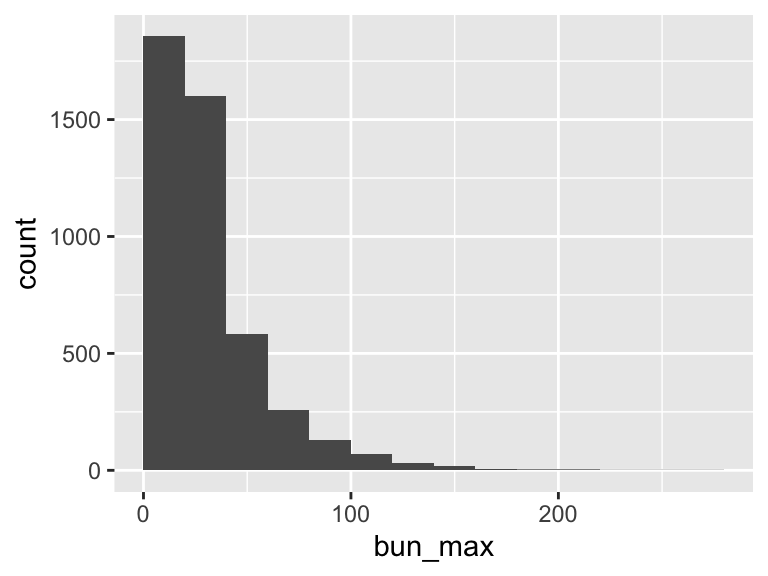
- Note that the following possible outlier values were detected: "3", "4", "5", "6", "7", …, "212", "218", "229", "251", "261" (13 values omitted).
bun_mean
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 256 |
| Median | 21.5 |
| 1st and 3rd quartiles | 14.5; 36 |
| Min. and max. | 0; 216.5 |
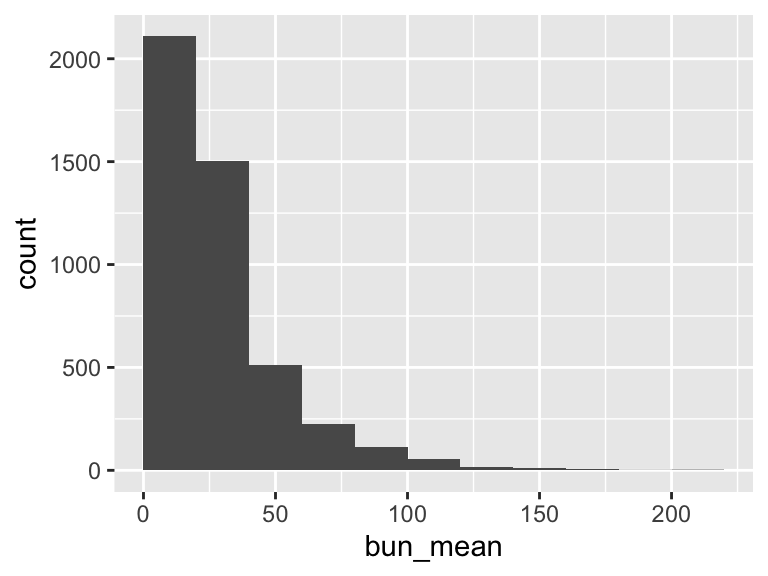
- Note that the following possible outlier values were detected: "0", "2", "3", "3.5", "4", …, "172", "178.5", "194.5", "206.5", "216.5" (19 values omitted).
wbc_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 5 (0.11 %) |
| Number of unique values | 348 |
| Median | 9.6 |
| 1st and 3rd quartiles | 6.6; 13.4 |
| Min. and max. | 0.1; 336.7 |
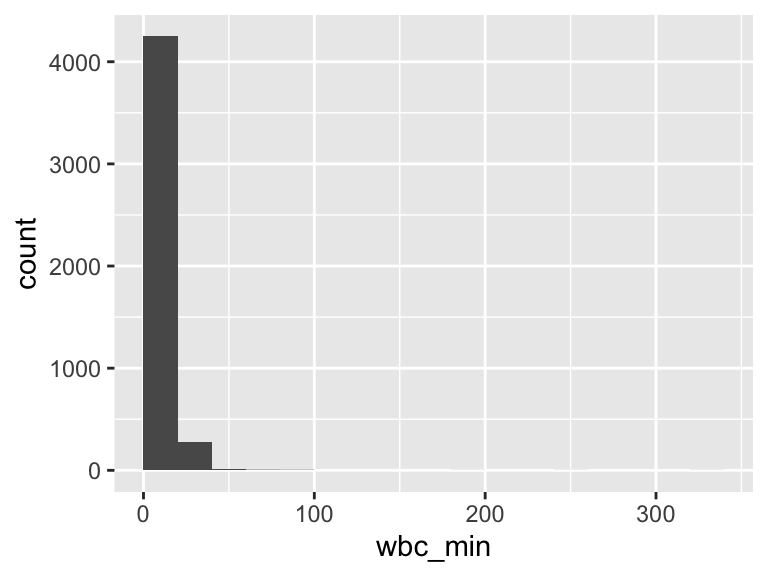
- Note that the following possible outlier values were detected: "0.1", "0.2", "0.3", "0.4", "0.5", …, "86", "87.8", "181.4", "243.6", "336.7" (66 values omitted).
wbc_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 5 (0.11 %) |
| Number of unique values | 435 |
| Median | 13.6 |
| 1st and 3rd quartiles | 9.7; 18.5 |
| Min. and max. | 0.1; 471.7 |
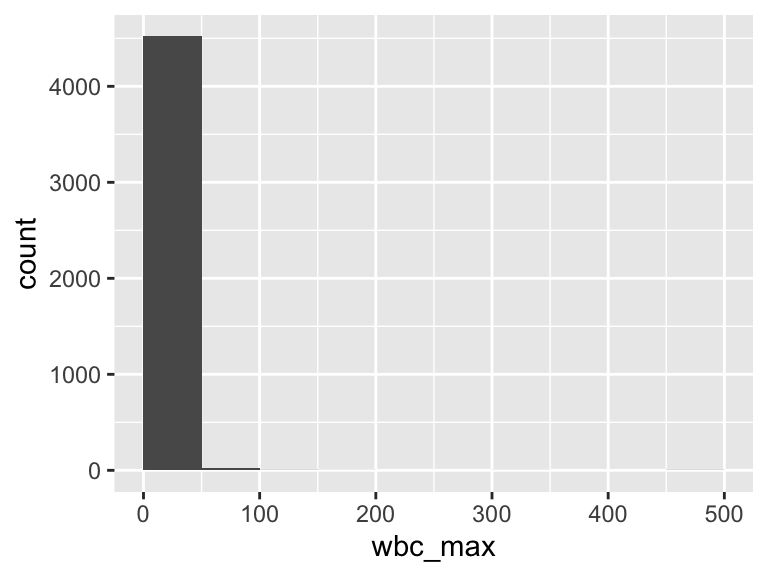
- Note that the following possible outlier values were detected: "0.1", "0.2", "0.3", "0.4", "0.5", …, "126.2", "183", "325.7", "462.6", "471.7" (78 values omitted).
wbc_mean
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 658 |
| Median | 11.75 |
| 1st and 3rd quartiles | 8.4; 15.85 |
| Min. and max. | 0; 404.2 |
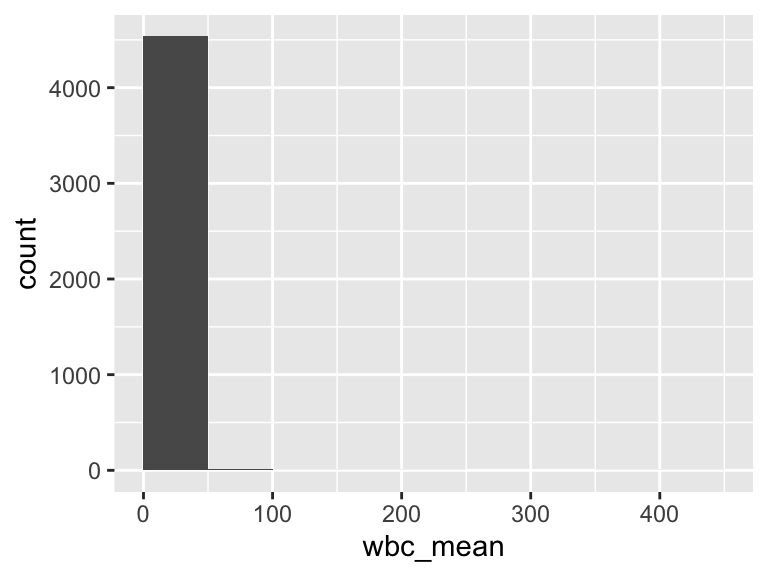
- Note that the following possible outlier values were detected: "0", "0.1", "0.15", "0.2", "0.25", …, "106.1", "182.2", "206.75", "353.1", "404.2" (94 values omitted).
heartrate_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 114 |
| Median | 72 |
| 1st and 3rd quartiles | 62; 84 |
| Min. and max. | 1; 134 |
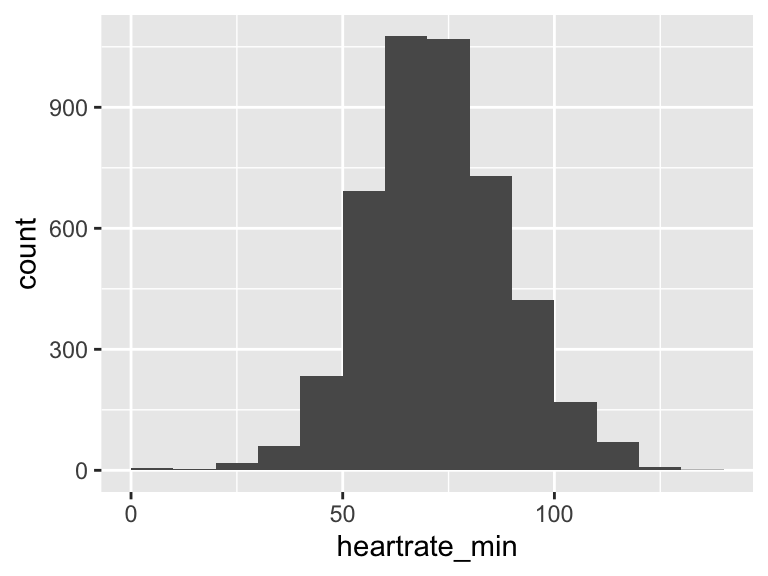
- Note that the following possible outlier values were detected: "1", "3", "4", "5", "8", …, "37", "127", "129", "130", "134" (17 values omitted).
heartrate_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 131 |
| Median | 106 |
| 1st and 3rd quartiles | 92; 121 |
| Min. and max. | 41; 223 |
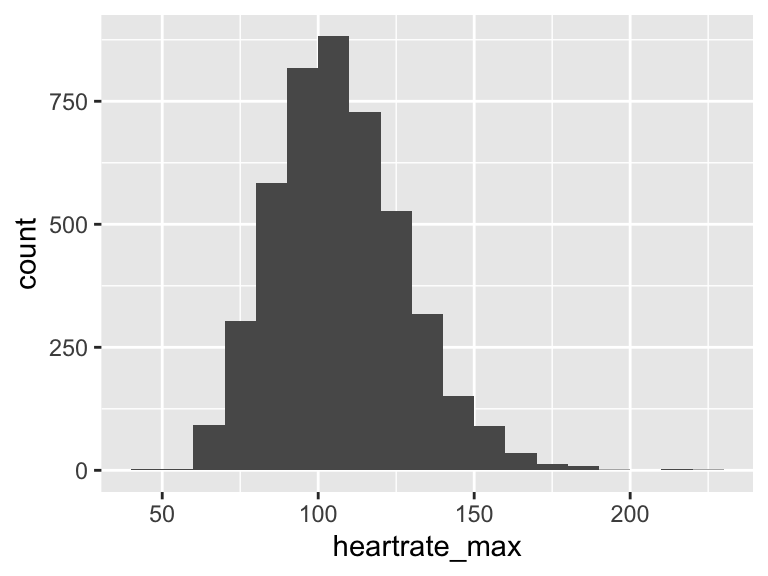
- Note that the following possible outlier values were detected: "41", "49", "50", "53", "55", …, "190", "194", "213", "215", "223" (9 values omitted).
heartrate_mean
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 3902 |
| Median | 87.78 |
| 1st and 3rd quartiles | 76.02; 99.75 |
| Min. and max. | 35.78; 154.53 |
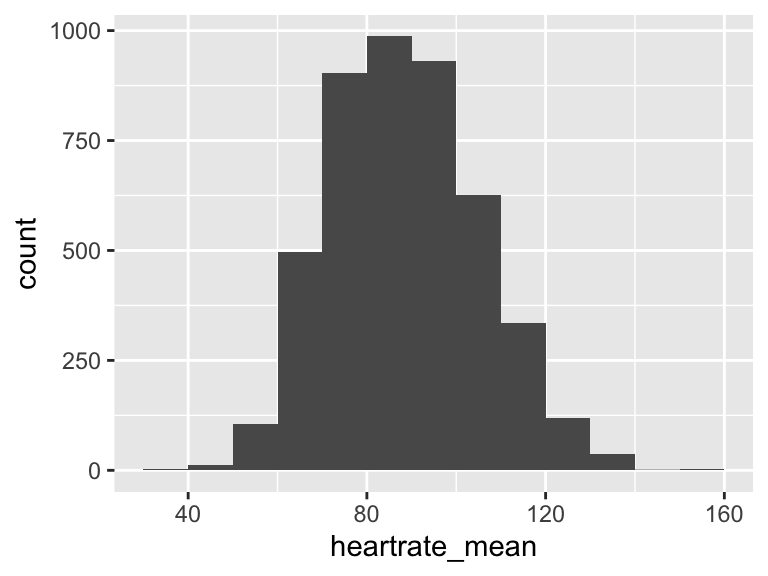
- Note that the following possible outlier values were detected: "35.78", "36", "37.62", "138.48", "138.71", …, "139.14", "139.48", "149.12", "152.22", "154.53" (1 values omitted).
sysbp_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 8 (0.18 %) |
| Number of unique values | 134 |
| Median | 88 |
| 1st and 3rd quartiles | 79; 98 |
| Min. and max. | 5; 168 |
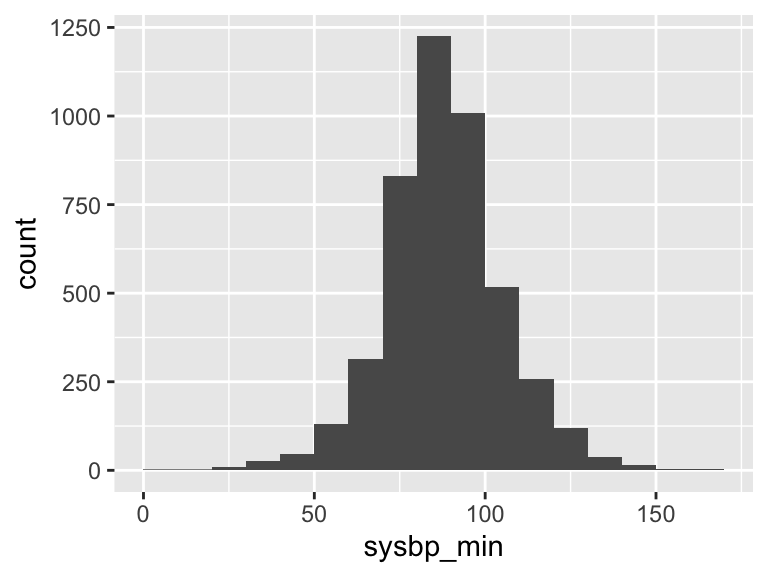
- Note that the following possible outlier values were detected: "5", "10", "18", "22", "23", …, "151", "158", "161", "162", "168" (49 values omitted).
sysbp_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 8 (0.18 %) |
| Number of unique values | 151 |
| Median | 146 |
| 1st and 3rd quartiles | 132; 162 |
| Min. and max. | 83; 311 |
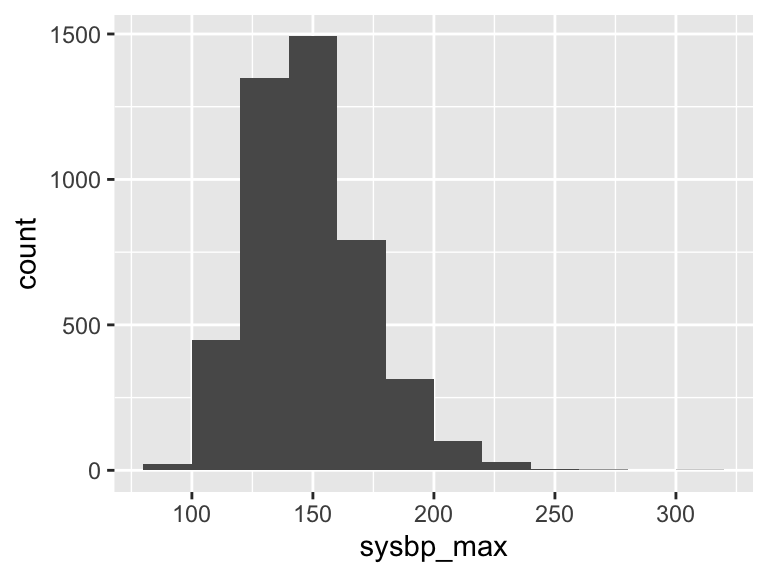
- Note that the following possible outlier values were detected: "83", "89", "90", "91", "92", …, "252", "254", "265", "273", "311" (21 values omitted).
sysbp_mean
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 8 (0.18 %) |
| Number of unique values | 3867 |
| Median | 113.61 |
| 1st and 3rd quartiles | 104.89; 125.63 |
| Min. and max. | 69.88; 195.34 |
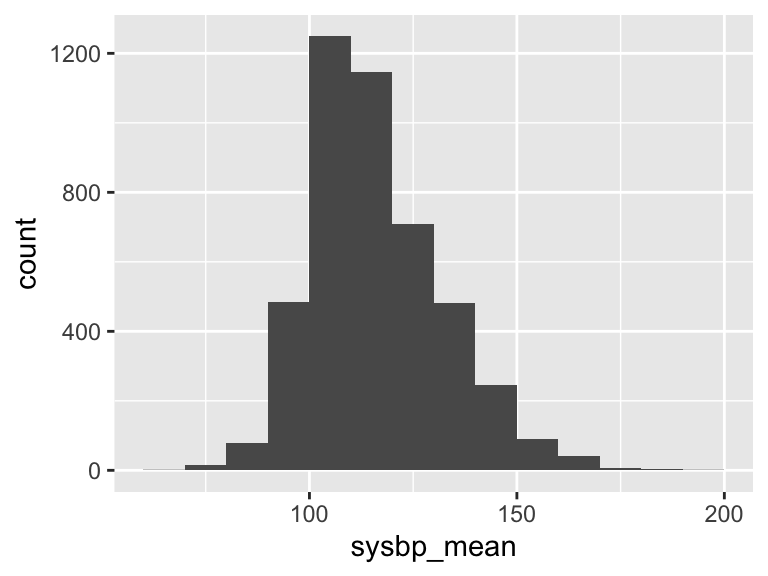
- Note that the following possible outlier values were detected: "69.88", "71.68", "71.73", "73.12", "75.44", …, "180.83", "181.96", "182.48", "188.29", "195.34" (85 values omitted).
diasbp_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 8 (0.18 %) |
| Number of unique values | 80 |
| Median | 44 |
| 1st and 3rd quartiles | 37; 51 |
| Min. and max. | 5; 114 |
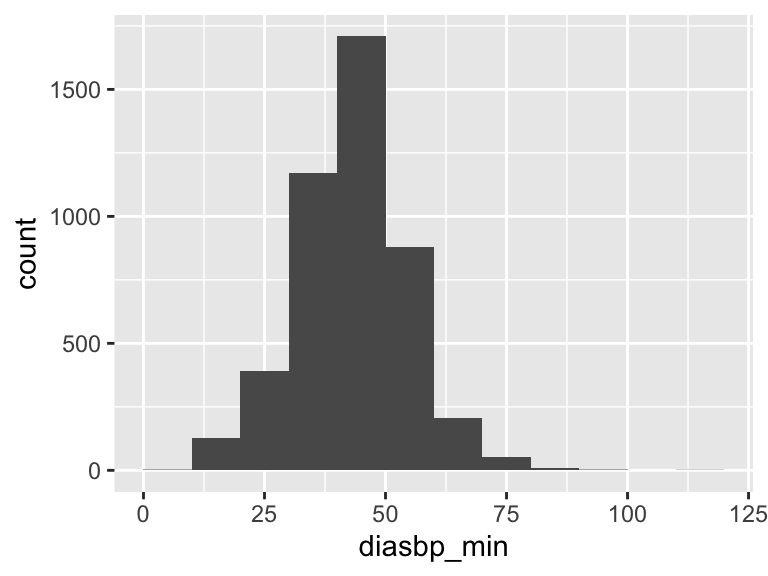
- Note that the following possible outlier values were detected: "5", "6", "11", "12", "13", …, "85", "88", "92", "93", "114" (13 values omitted).
diasbp_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 8 (0.18 %) |
| Number of unique values | 136 |
| Median | 83 |
| 1st and 3rd quartiles | 73; 96 |
| Min. and max. | 35; 197 |
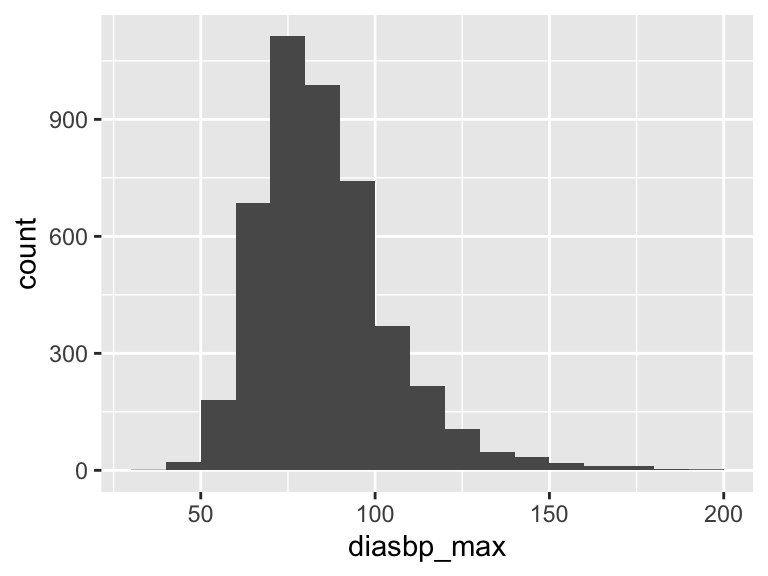
- Note that the following possible outlier values were detected: "35", "41", "44", "46", "48", …, "183", "185", "187", "196", "197" (30 values omitted).
diasbp_mean
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 8 (0.18 %) |
| Number of unique values | 3621 |
| Median | 60.3 |
| 1st and 3rd quartiles | 54.09; 67 |
| Min. and max. | 26.33; 129.57 |
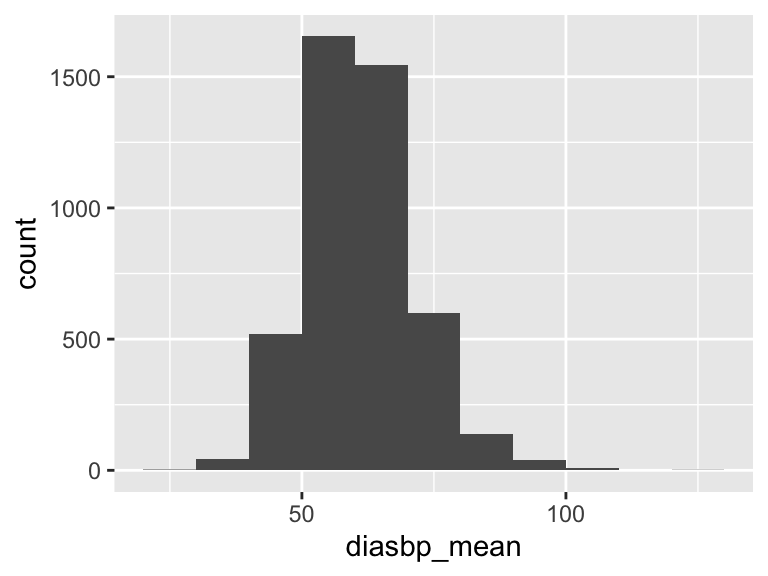
- Note that the following possible outlier values were detected: "26.33", "28.4", "32.48", "32.94", "33.43", …, "103.52", "104.29", "105.46", "107.43", "129.57" (67 values omitted).
meanbp_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 103 |
| Median | 56 |
| 1st and 3rd quartiles | 49; 63 |
| Min. and max. | 1; 113 |
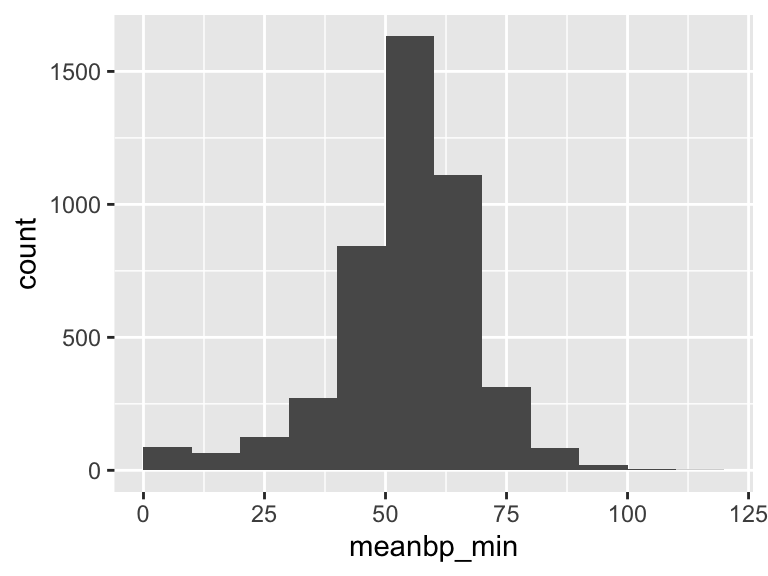
- Note that the following possible outlier values were detected: "1", "2", "3", "4", "5", …, "101", "102", "104", "109", "113" (37 values omitted).
meanbp_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 197 |
| Median | 101 |
| 1st and 3rd quartiles | 90; 114 |
| Min. and max. | 56; 298 |
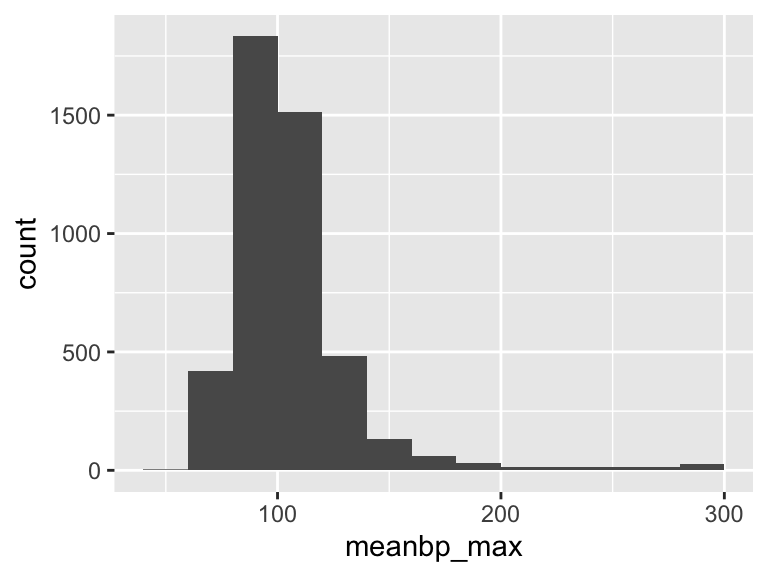
- Note that the following possible outlier values were detected: "56", "58", "60", "62", "63", …, "294", "295", "296", "297", "298" (89 values omitted).
meanbp_mean
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 3677 |
| Median | 75 |
| 1st and 3rd quartiles | 69.04; 82.41 |
| Min. and max. | 45.6; 133.16 |
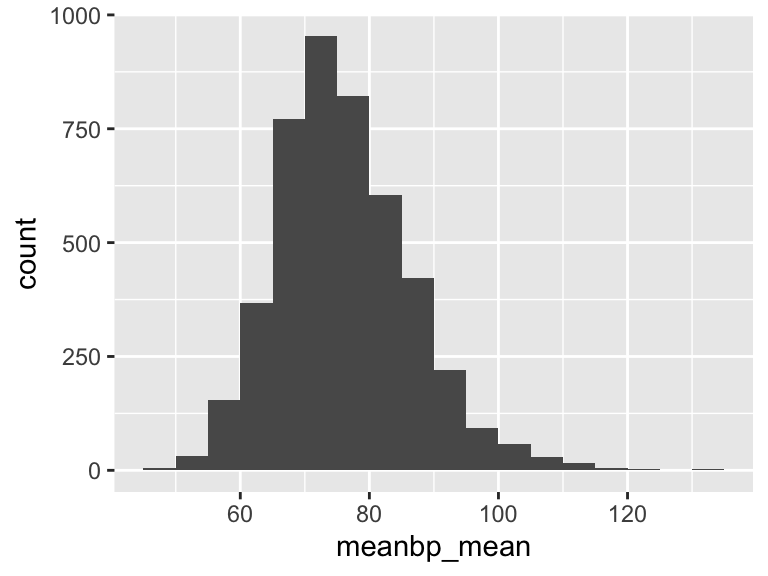
- Note that the following possible outlier values were detected: "45.6", "46.97", "48.36", "48.71", "49.83", …, "122.48", "123.21", "124.96", "131.5", "133.16" (71 values omitted).
resprate_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 1 (0.02 %) |
| Number of unique values | 30 |
| Median | 13 |
| 1st and 3rd quartiles | 10; 15 |
| Min. and max. | 1; 32 |
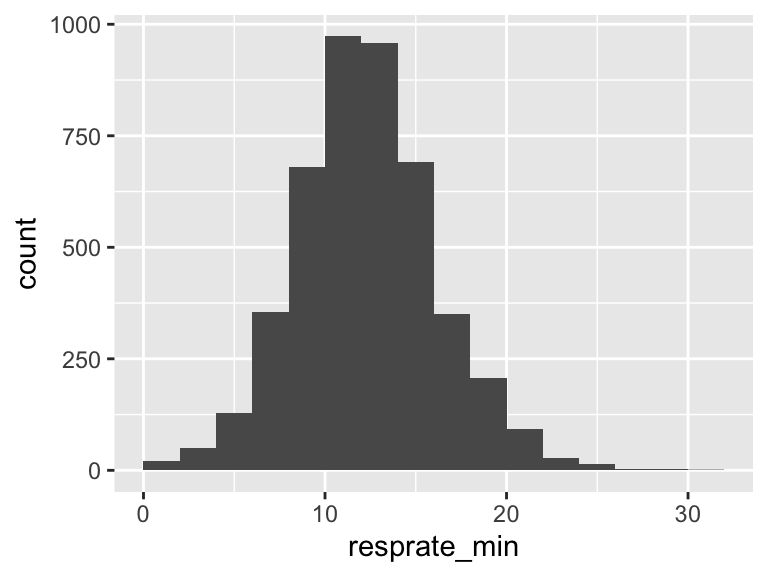
- Note that the following possible outlier values were detected: "1", "2", "23", "24", "25", "26", "27", "28", "30", "32".
resprate_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 1 (0.02 %) |
| Number of unique values | 53 |
| Median | 27 |
| 1st and 3rd quartiles | 24; 32 |
| Min. and max. | 13; 69 |
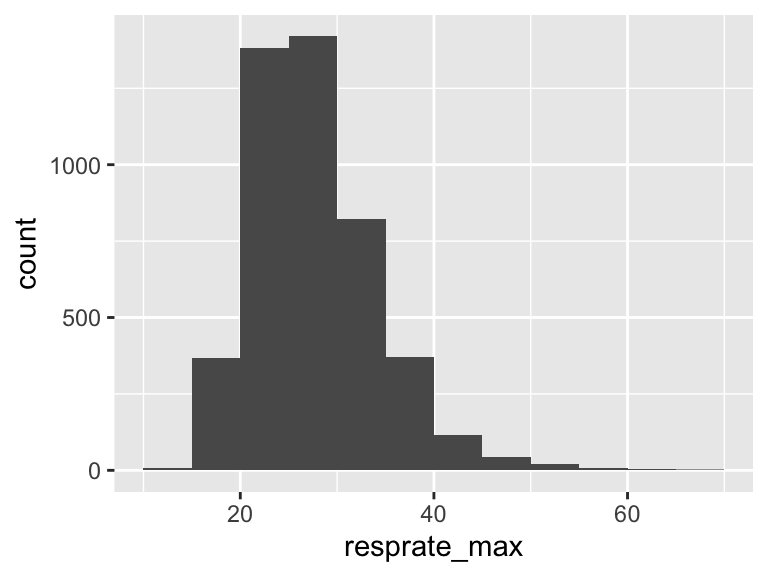
- Note that the following possible outlier values were detected: "13", "14", "15", "16", "17", …, "61", "62", "64", "68", "69" (10 values omitted).
resprate_mean
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 1 (0.02 %) |
| Number of unique values | 3110 |
| Median | 19.26 |
| 1st and 3rd quartiles | 16.79; 22.4 |
| Min. and max. | 9.54; 40.58 |
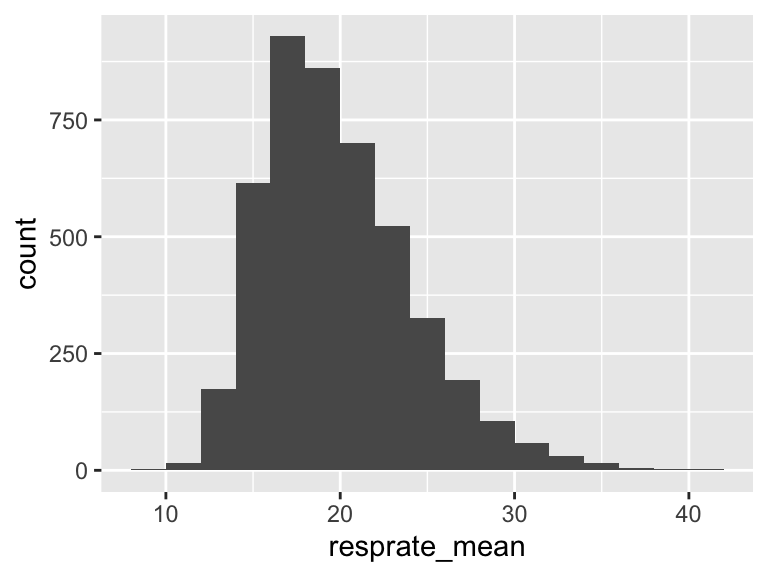
- Note that the following possible outlier values were detected: "9.54", "9.78", "10.17", "10.5", "10.79", …, "38.44", "38.96", "40.16", "40.37", "40.58" (20 values omitted).
tempc_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 103 (2.26 %) |
| Number of unique values | 167 |
| Median | 36.11 |
| 1st and 3rd quartiles | 35.61; 36.61 |
| Min. and max. | 15; 39.6 |
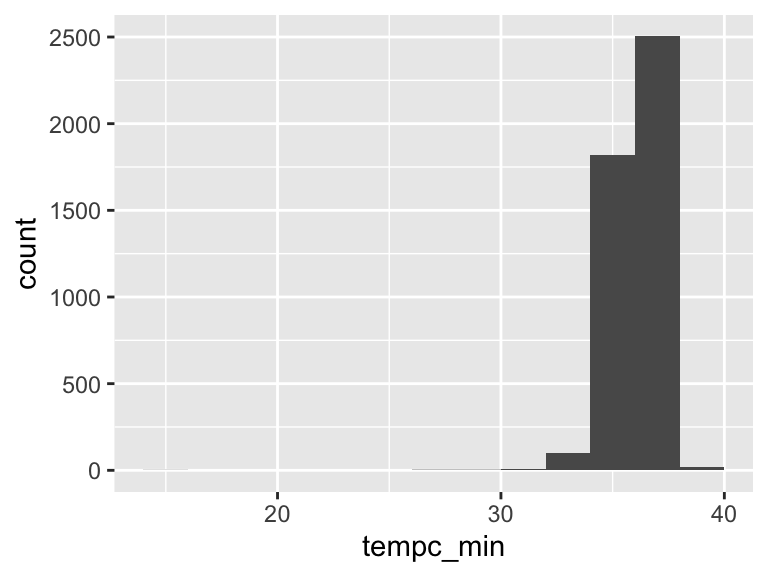
- Note that the following possible outlier values were detected: "15", "26.67", "29.44", "29.9", "30.56", …, "38.7", "38.72", "38.94", "39.2", "39.6" (53 values omitted).
tempc_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 103 (2.26 %) |
| Number of unique values | 164 |
| Median | 37.5 |
| 1st and 3rd quartiles | 37; 38.11 |
| Min. and max. | 31.6; 42 |
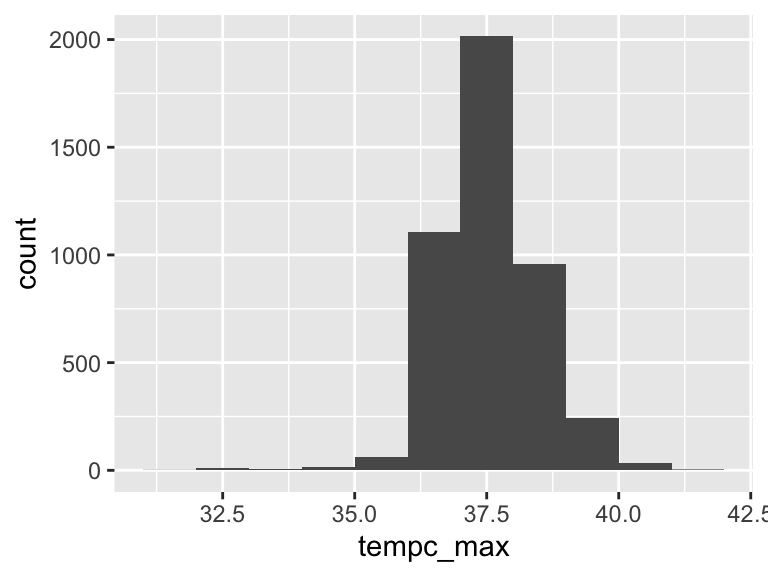
- Note that the following possible outlier values were detected: "31.6", "32.2", "32.4", "32.61", "32.7", …, "40.9", "40.94", "41.11", "41.6", "42" (39 values omitted).
tempc_mean
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 103 (2.26 %) |
| Number of unique values | 1692 |
| Median | 36.82 |
| 1st and 3rd quartiles | 36.41; 37.31 |
| Min. and max. | 31.6; 40.1 |
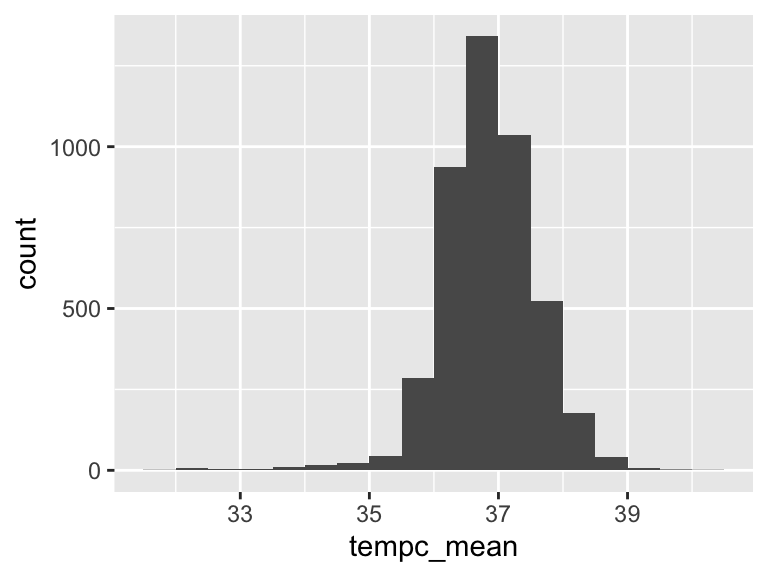
- Note that the following possible outlier values were detected: "31.6", "32.02", "32.1", "32.2", "32.35", …, "39.36", "39.45", "39.56", "39.71", "40.1" (84 values omitted).
spo2_min
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 1 (0.02 %) |
| Number of unique values | 71 |
| Median | 93 |
| 1st and 3rd quartiles | 90; 95 |
| Min. and max. | 1; 100 |
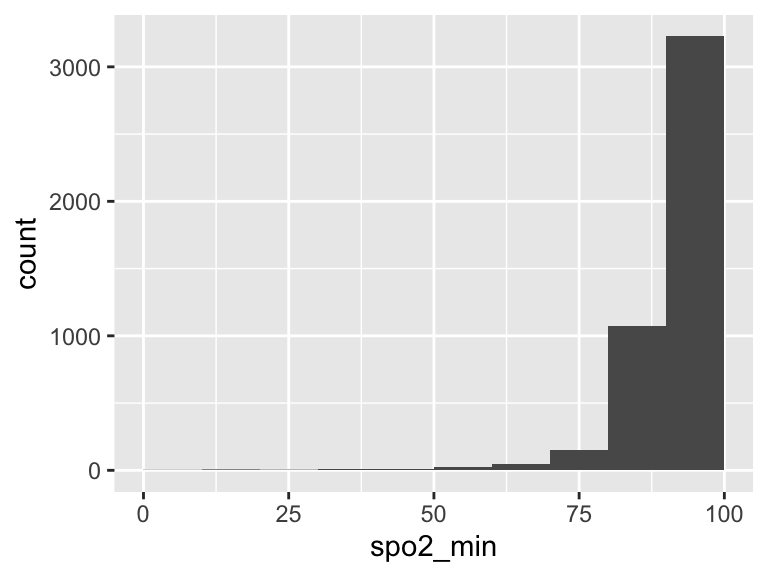
- Note that the following possible outlier values were detected: "1", "5", "11", "13", "18", …, "74", "75", "76", "99", "100" (39 values omitted).
spo2_max
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 1 (0.02 %) |
| Number of unique values | 12 |
| Median | 100 |
| 1st and 3rd quartiles | 100; 100 |
| Min. and max. | 89; 100 |
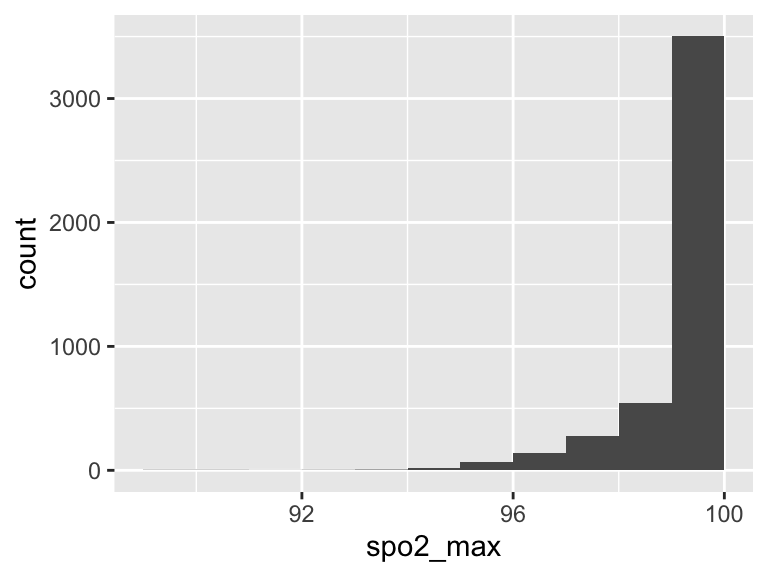
- Note that the following possible outlier values were detected: "89", "90", "91", "92", "93", …, "95", "96", "97", "98", "99" (1 values omitted).
spo2_mean
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 1 (0.02 %) |
| Number of unique values | 2015 |
| Median | 97.26 |
| 1st and 3rd quartiles | 95.8; 98.6 |
| Min. and max. | 55.87; 100 |
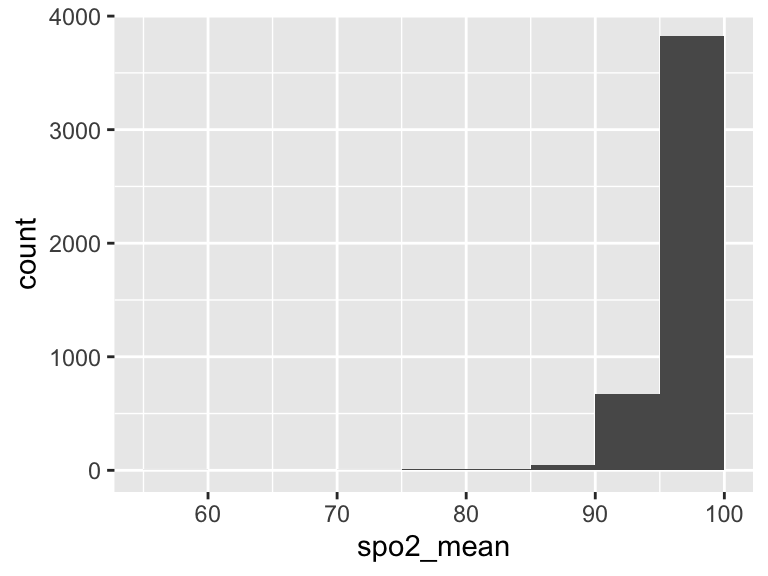
- Note that the following possible outlier values were detected: "55.87", "71.85", "76.16", "76.38", "78.08", …, "89.81", "89.96", "90", "90.07", "90.13" (55 values omitted).
glucose_min1
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 30 (0.66 %) |
| Number of unique values | 245 |
| Median | 106 |
| 1st and 3rd quartiles | 89; 127 |
| Min. and max. | 12; 480 |
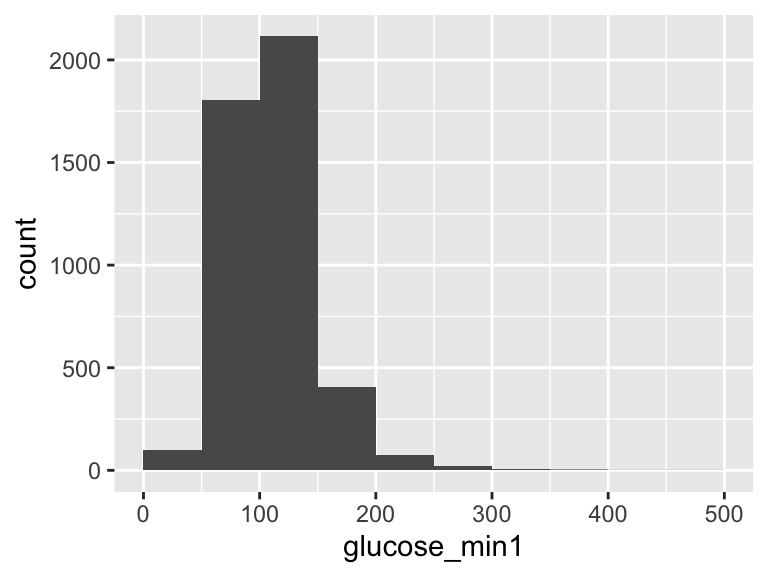
- Note that the following possible outlier values were detected: "12", "15", "16", "19", "20", …, "356", "357", "374", "424", "480" (80 values omitted).
glucose_max1
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 30 (0.66 %) |
| Number of unique values | 454 |
| Median | 166 |
| 1st and 3rd quartiles | 130; 217 |
| Min. and max. | 57; 999999 |
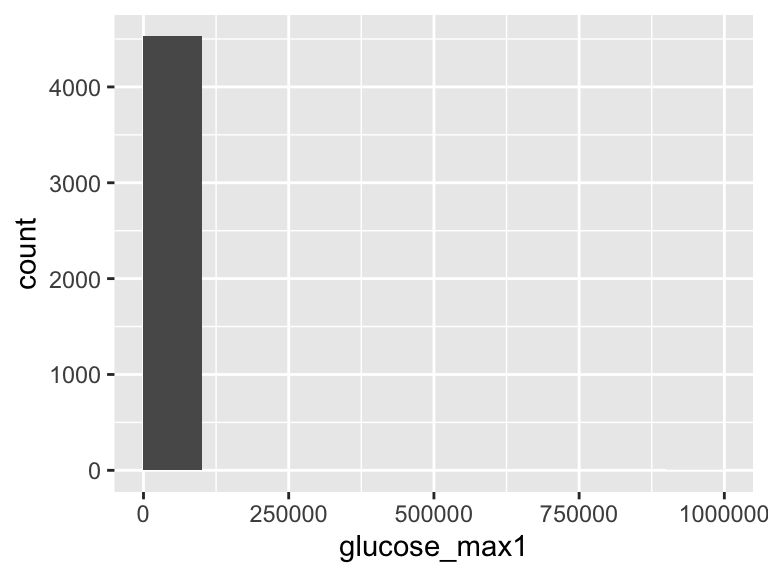
The following suspected missing value codes enter as regular values: "999999".
Note that the following possible outlier values were detected: "57", "62", "64", "65", "66", …, "883", "900", "1016", "1038", "999999" (62 values omitted).
glucose_mean
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 30 (0.66 %) |
| Number of unique values | 2228 |
| Median | 134.19 |
| 1st and 3rd quartiles | 113; 165 |
| Min. and max. | 52.44; 142966.86 |
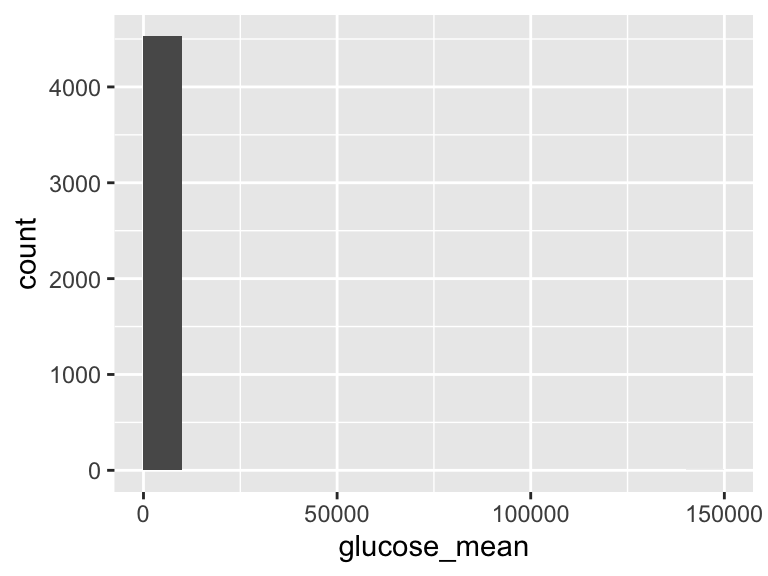
- Note that the following possible outlier values were detected: "52.44", "57", "62", "62.75", "63.07", …, "479.26", "578.25", "657", "786.22", "142966.86" (114 values omitted).
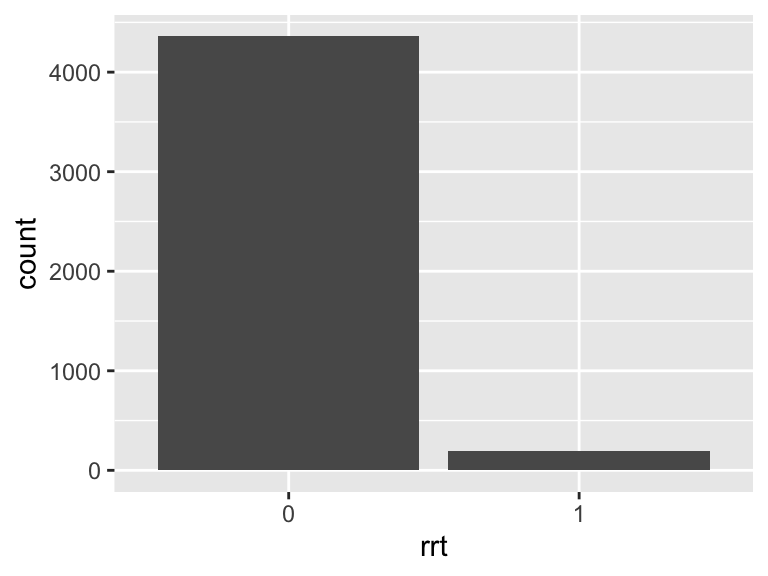
rrt
- Note that this variable is treated as a factor variable below, as it only takes a few unique values.
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 2 |
| Mode | “0” |
| Reference category | 0 |
subject_id
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 4559 |
| Median | 68368 |
| 1st and 3rd quartiles | 53111.5; 83771.5 |
| Min. and max. | 165; 99982 |
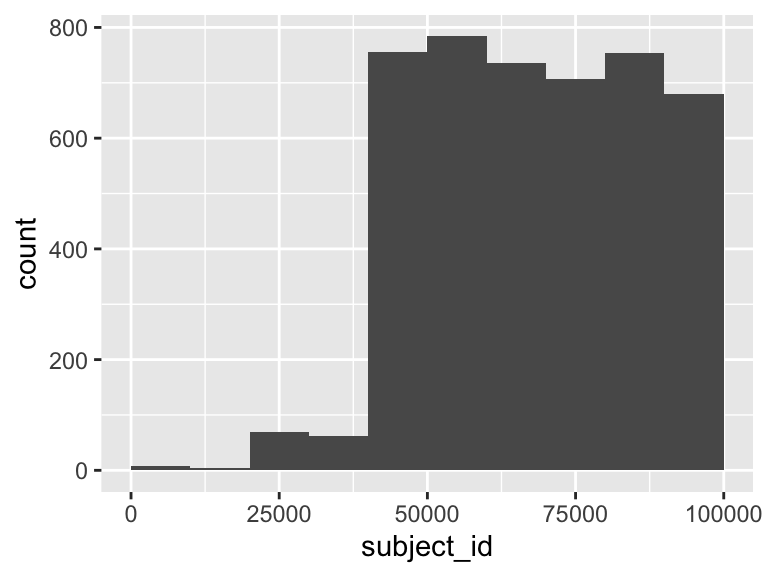
- Note that the following possible outlier values were detected: "165", "266", "671", "5771", "7310".
hadm_id…102
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 4559 |
| Median | 149643 |
| 1st and 3rd quartiles | 125389; 175033 |
| Min. and max. | 100003; 199962 |
icustay_id…103
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 4559 |
| Median | 251008 |
| 1st and 3rd quartiles | 225575.5; 275526 |
| Min. and max. | 200075; 299998 |
urineoutput
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 0 (0 %) |
| Number of unique values | 1794 |
| Median | 1560 |
| 1st and 3rd quartiles | 897.5; 2460 |
| Min. and max. | 0; 50515 |
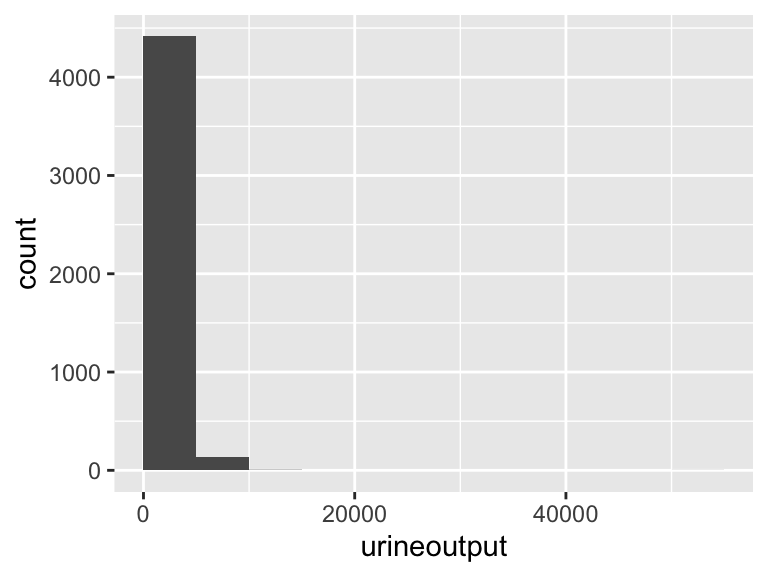
- Note that the following possible outlier values were detected: "6540", "6560", "6640", "6650", "6670", …, "9910", "10525", "11025", "12210", "50515" (34 values omitted).
colloid_bolus
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 4050 (88.84 %) |
| Number of unique values | 13 |
| Median | 500 |
| 1st and 3rd quartiles | 250; 500 |
| Min. and max. | 150; 1000 |
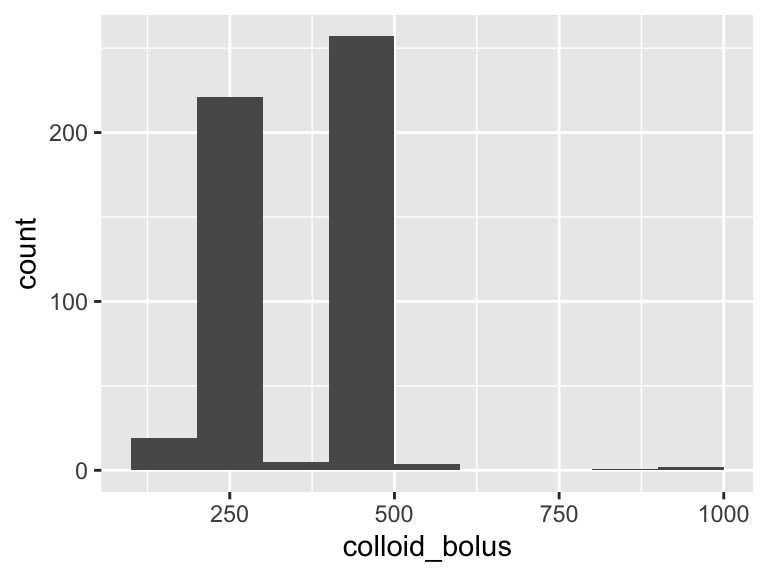
- Note that the following possible outlier values were detected: "550", "600", "900", "1000".
crystalloid_bolus
| Feature | Result |
|---|---|
| Variable type | numeric |
| Number of missing obs. | 1194 (26.19 %) |
| Number of unique values | 96 |
| Median | 500 |
| 1st and 3rd quartiles | 500; 1000 |
| Min. and max. | 250; 11000 |
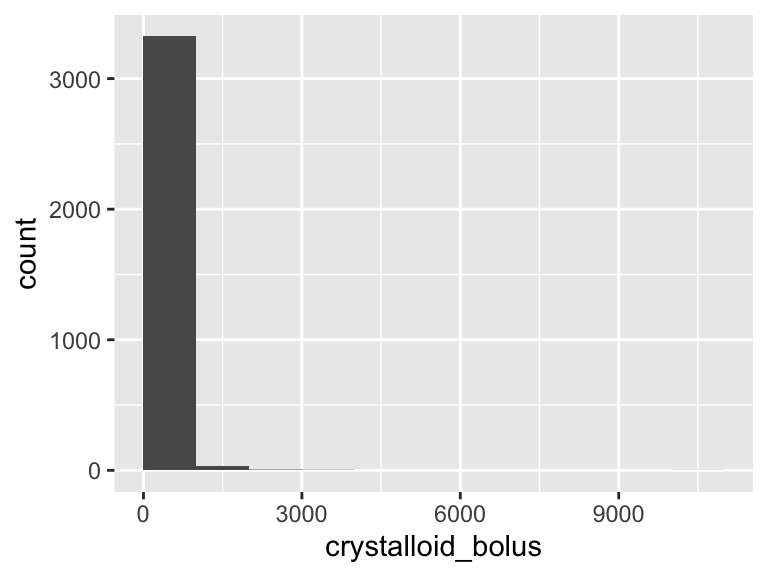
- Note that the following possible outlier values were detected: "250", "258", "267", "275", "276", …, "423", "433", "450", "467", "475" (21 values omitted).
Report generation information:
Created by: Could not determine from system (username:
ecm20).Report creation time: Sun Oct 08 2023 10:44:20
Report was run from directory:
/home/ecm20/R/STA6257_Project_SVM/Josh Branch/STA6257_Project_SVM_Josh/Team RepodataMaid v1.4.1 [Pkg: 2021-10-08 from CRAN (R 4.3.1)]
R version 4.3.1 (2023-06-16).
Platform: x86_64-pc-linux-gnu (64-bit)(America/Chicago).
Function call:
makeDataReport(data = data, output = "html", replace = TRUE)